Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ
- PMID: 32276965
- PMCID: PMC7296464
- DOI: 10.1126/sciimmunol.aax0062
Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ
Abstract
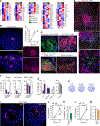
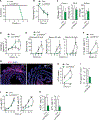
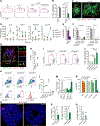
Intestinal mononuclear phagocytes (MPs) are composed of heterogeneous dendritic cell (DC) and macrophage subsets necessary for the initiation of immune response and control of inflammation. Although MPs in the normal intestine have been extensively studied, the heterogeneity and function of inflammatory MPs remain poorly defined. We performed phenotypical, transcriptional, and functional analyses of inflammatory MPs in infectious Salmonella colitis and identified CX3CR1+ MPs as the most prevalent inflammatory cell type. CX3CR1+ MPs were further divided into three distinct populations, namely, Nos2 +CX3CR1lo, Ccr7 +CX3CR1int (lymph migratory), and Cxcl13 +CX3CR1hi (mucosa resident), all of which were transcriptionally aligned with macrophages and derived from monocytes. In follow-up experiments in vivo, intestinal CX3CR1+ macrophages were superior to conventional DC1 (cDC1) and cDC2 in inducing Salmonella-specific mucosal IgA. We next examined spatial organization of the immune response induced by CX3CR1+ macrophage subsets and identified mucosa-resident Cxcl13 +CX3CR1hi macrophages as the antigen-presenting cells responsible for recruitment and activation of CD4+ T and B cells to the sites of Salmonella invasion, followed by tertiary lymphoid structure formation and the local pathogen-specific IgA response. Using mice we developed with a floxed Ccr7 allele, we showed that this local IgA response developed independently of migration of the Ccr7 +CX3CR1int population to the mesenteric lymph nodes and contributed to the total mucosal IgA response to infection. The differential activity of intestinal macrophage subsets in promoting mucosal IgA responses should be considered in the development of vaccines to prevent Salmonella infection and in the design of anti-inflammatory therapies aimed at modulating macrophage function in inflammatory bowel disease.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




Similar articles
-
CX3CR1+ Macrophages and CD8+ T Cells Control Intestinal IgA Production.J Immunol. 2018 Aug 15;201(4):1287-1294. doi: 10.4049/jimmunol.1701459. Epub 2018 Jul 9. J Immunol. 2018. PMID: 29987162 Free PMC article.
-
Saccharomyces boulardii Strain CNCM I-745 Modifies the Mononuclear Phagocytes Response in the Small Intestine of Mice Following Salmonella Typhimurium Infection.Front Immunol. 2019 Apr 2;10:643. doi: 10.3389/fimmu.2019.00643. eCollection 2019. Front Immunol. 2019. PMID: 31001263 Free PMC article.
-
Dynamic role of macrophage CX3CR1 expression in inflammatory bowel disease.Immunol Lett. 2021 Apr;232:39-44. doi: 10.1016/j.imlet.2021.02.001. Epub 2021 Feb 11. Immunol Lett. 2021. PMID: 33582183 Review.
-
CX3CR1 defines functionally distinct intestinal mononuclear phagocyte subsets which maintain their respective functions during homeostatic and inflammatory conditions.Eur J Immunol. 2011 Mar;41(3):773-9. doi: 10.1002/eji.201040965. Epub 2011 Feb 11. Eur J Immunol. 2011. PMID: 21341263
-
Re-thinking the functions of IgA(+) plasma cells.Gut Microbes. 2014;5(5):652-62. doi: 10.4161/19490976.2014.969977. Gut Microbes. 2014. PMID: 25483334 Free PMC article. Review.
Cited by
-
Emerging concepts in intestinal immune control of obesity-related metabolic disease.Nat Commun. 2021 May 10;12(1):2598. doi: 10.1038/s41467-021-22727-7. Nat Commun. 2021. PMID: 33972511 Free PMC article. Review.
-
Microbial energy metabolism fuels an intestinal macrophage niche in solitary isolated lymphoid tissues through purinergic signaling.Sci Immunol. 2023 Aug 4;8(86):eabq4573. doi: 10.1126/sciimmunol.abq4573. Epub 2023 Aug 4. Sci Immunol. 2023. PMID: 37540734 Free PMC article.
-
Tertiary lymphoid structures and B lymphocytes: a promising therapeutic strategy to fight cancer.Front Immunol. 2023 Aug 9;14:1231315. doi: 10.3389/fimmu.2023.1231315. eCollection 2023. Front Immunol. 2023. PMID: 37622111 Free PMC article. Review.
-
Macrophages in intestinal homeostasis and inflammatory bowel disease.Nat Rev Gastroenterol Hepatol. 2023 Aug;20(8):538-553. doi: 10.1038/s41575-023-00769-0. Epub 2023 Apr 17. Nat Rev Gastroenterol Hepatol. 2023. PMID: 37069320 Review.
-
Contribution of macrophages to neural survival and intracochlear tissue remodeling responses following cochlear implantation.J Neuroinflammation. 2023 Nov 16;20(1):266. doi: 10.1186/s12974-023-02955-y. J Neuroinflammation. 2023. PMID: 37974203 Free PMC article.
References
-
- Joeris T, Müller-Luda K, Agace WW, Mowat AM, Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. 10, 845–864 (2017). - PubMed
-
- Bain CC, Montgomery J, Scott CL, Kel JM, Girard-Madoux MJH, Martens L, Zangerle-Murray TFP, Ober-Blöbaum J, Lindenbergh-Kortleve D, Samsom JN, Henri S, Lawrence T, Saeys Y, Malissen B, Dalod M, Clausen BE, Mowat AM, TGFβR signalling controls CD103+CD11b+ dendritic cell development in the intestine. Nat. Commun 8, 620 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

