A Two-Component regulatory system with opposite effects on glycopeptide antibiotic biosynthesis and resistance
- PMID: 32277112
- PMCID: PMC7148328
- DOI: 10.1038/s41598-020-63257-4
A Two-Component regulatory system with opposite effects on glycopeptide antibiotic biosynthesis and resistance
Abstract
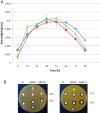
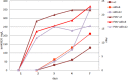
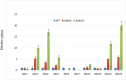
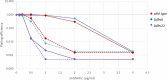
The glycopeptide A40926, produced by the actinomycete Nonomuraea gerenzanensis, is the precursor of dalbavancin, a second-generation glycopeptide antibiotic approved for clinical use in the USA and Europe in 2014 and 2015, respectively. The final product of the biosynthetic pathway is an O-acetylated form of A40926 (acA40926). Glycopeptide biosynthesis in N. gerenzanensis is dependent upon the dbv gene cluster that encodes, in addition to the two essential positive regulators Dbv3 and Dbv4, the putative members of a two-component signal transduction system, specifically the response regulator Dbv6 and the sensor kinase Dbv22. The aim of this work was to assign a role to these two genes. Our results demonstrate that deletion of dbv22 leads to an increased antibiotic production with a concomitant reduction in glycopeptide resistance. Deletion of dbv6 results in a similar phenotype, although the effects are not as strong as in the Δdbv22 mutant. Consistently, quantitative RT-PCR analysis showed that Dbv6 and Dbv22 negatively regulate the regulatory genes (dbv3 and dbv4), as well as some dbv biosynthetic genes (dbv23 and dbv24), whereas Dbv6 and Dbv22 positively regulate transcription of the single, cluster-associated resistance gene. Finally, we demonstrate that exogenously added acA40926 and its precursor A40926 can modulate transcription of dbv genes but with an opposite extent: A40926 strongly stimulates transcription of the Dbv6/Dbv22 target genes while acA40926 has a neutral or negative effect on transcription of those genes. We propose a model in which glycopeptide biosynthesis in N. gerenzanensis is modulated through a positive feedback by the biosynthetic precursor A40926 and a negative feedback by the final product acA40926. In addition to previously reported control systems, this sophisticated control loop might help the producing strain cope with the toxicity of its own product. This work, besides leading to improved glycopeptide producing strains, enlarges our knowledge on the regulation of glycopeptide biosynthesis in actinomycetes, setting N. gerenzanensis and its two-component system Dbv6-Dbv22 apart from other glycopeptide producers.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

