BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy
- PMID: 32277346
- PMCID: PMC7452999
- DOI: 10.1007/s13238-020-00713-x
BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy
Abstract
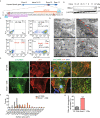
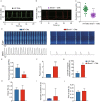
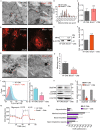
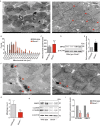
Dysregulation of circadian rhythms associates with cardiovascular disorders. It is known that deletion of the core circadian gene Bmal1 in mice causes dilated cardiomyopathy. However, the biological rhythm regulation system in mouse is very different from that of humans. Whether BMAL1 plays a role in regulating human heart function remains unclear. Here we generated a BMAL1 knockout human embryonic stem cell (hESC) model and further derived human BMAL1 deficient cardiomyocytes. We show that BMAL1 deficient hESC-derived cardiomyocytes exhibited typical phenotypes of dilated cardiomyopathy including attenuated contractility, calcium dysregulation, and disorganized myofilaments. In addition, mitochondrial fission and mitophagy were suppressed in BMAL1 deficient hESC-cardiomyocytes, which resulted in significantly attenuated mitochondrial oxidative phosphorylation and compromised cardiomyocyte function. We also found that BMAL1 binds to the E-box element in the promoter region of BNIP3 gene and specifically controls BNIP3 protein expression. BMAL1 knockout directly reduced BNIP3 protein level, causing compromised mitophagy and mitochondria dysfunction and thereby leading to compromised cardiomyocyte function. Our data indicated that the core circadian gene BMAL1 is critical for normal mitochondria activities and cardiac function. Circadian rhythm disruption may directly link to compromised heart function and dilated cardiomyopathy in humans.
Keywords: cardiomyocytes; cell differentiation; circadian gene BMAL1; dilated cardiomyopathy; human embryonic stem cells; mitochondria.
Figures




References
-
- Alibhai FJ, LaMarre J, Reitz CJ, Tsimakouridze EV, Kroetsch JT, Bolz SS, Shulman A, Steinberg S, Burris TP, Oudit GY, et al. Disrupting the key circadian regulator CLOCK leads to age-dependent cardiovascular disease. J Mol Cell Cardiol. 2017;105:24–37. - PubMed
-
- Alibhai FJ, Reitz CJ, Peppler WT, Basu P, Sheppard P, Choleris E, Bakovic M, Martino TA. Female ClockDelta19/Delta19 mice are protected from the development of age-dependent cardiomyopathy. Cardiovasc Res. 2018;114:259–271. - PubMed
-
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

