Determinants of cyclization-decyclization kinetics of short DNA with sticky ends
- PMID: 32282905
- PMCID: PMC7229855
- DOI: 10.1093/nar/gkaa207
Determinants of cyclization-decyclization kinetics of short DNA with sticky ends
Abstract
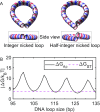
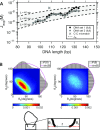
Cyclization of DNA with sticky ends is commonly used to measure DNA bendability as a function of length and sequence, but how its kinetics depend on the rotational positioning of the sticky ends around the helical axis is less clear. Here, we measured cyclization (looping) and decyclization (unlooping) rates (kloop and kunloop) of DNA with sticky ends over three helical periods (100-130 bp) using single-molecule fluorescence resonance energy transfer (FRET). kloop showed a nontrivial undulation as a function of DNA length whereas kunloop showed a clear oscillation with a period close to the helical turn of DNA (∼10.5 bp). The oscillation of kunloop was almost completely suppressed in the presence of gaps around the sticky ends. We explain these findings by modeling double-helical DNA as a twisted wormlike chain with a finite width, intrinsic curvature, and stacking interaction between the end base pairs. We also discuss technical issues for converting the FRET-based cyclization/decyclization rates to an equilibrium quantity known as the J factor that is widely used to characterize DNA bending mechanics.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Tardin C. The mechanics of DNA loops bridged by proteins unveiled by single-molecule experiments. Biochimie. 2017; 142:80–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

