The T1D-associated lncRNA Lnc13 modulates human pancreatic β cell inflammation by allele-specific stabilization of STAT1 mRNA
- PMID: 32284404
- PMCID: PMC7183221
- DOI: 10.1073/pnas.1914353117
The T1D-associated lncRNA Lnc13 modulates human pancreatic β cell inflammation by allele-specific stabilization of STAT1 mRNA
Abstract
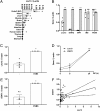
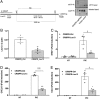
The vast majority of type 1 diabetes (T1D) genetic association signals lie in noncoding regions of the human genome. Many have been predicted to affect the expression and secondary structure of long noncoding RNAs (lncRNAs), but the contribution of these lncRNAs to the pathogenesis of T1D remains to be clarified. Here, we performed a complete functional characterization of a lncRNA that harbors a single nucleotide polymorphism (SNP) associated with T1D, namely, Lnc13 Human pancreatic islets harboring the T1D-associated SNP risk genotype in Lnc13 (rs917997*CC) showed higher STAT1 expression than islets harboring the heterozygous genotype (rs917997*CT). Up-regulation of Lnc13 in pancreatic β-cells increased activation of the proinflammatory STAT1 pathway, which correlated with increased production of chemokines in an allele-specific manner. In a mirror image, Lnc13 gene disruption in β-cells partially counteracts polyinosinic-polycytidylic acid (PIC)-induced STAT1 and proinflammatory chemokine expression. Furthermore, we observed that PIC, a viral mimetic, induces Lnc13 translocation from the nucleus to the cytoplasm promoting the interaction of STAT1 mRNA with (poly[rC] binding protein 2) (PCBP2). Interestingly, Lnc13-PCBP2 interaction regulates the stability of the STAT1 mRNA, sustaining inflammation in β-cells in an allele-specific manner. Our results show that the T1D-associated Lnc13 may contribute to the pathogenesis of T1D by increasing pancreatic β-cell inflammation. These findings provide information on the molecular mechanisms by which disease-associated SNPs in lncRNAs influence disease pathogenesis and open the door to the development of diagnostic and therapeutic approaches based on lncRNA targeting.
Keywords: inflammation; lncRNA; pancreatic β-cell; polymorphism; type 1 diabetes.
Conflict of interest statement
The authors declare no competing interest.
Figures




Comment in
-
Role of long non-coding RNA in T1DM.Nat Rev Endocrinol. 2020 Jul;16(7):344-345. doi: 10.1038/s41574-020-0368-2. Nat Rev Endocrinol. 2020. PMID: 32358540 No abstract available.
References
-
- Eizirik D. L., Colli M. L., Ortis F., The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 5, 219–226 (2009). - PubMed
-
- Santin I., Eizirik D. L., Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and β-cell apoptosis. Diabetes Obes. Metab. 15 (suppl. 3), 71–81 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

