Dynamic coupling of whole-brain neuronal and neurotransmitter systems
- PMID: 32284420
- PMCID: PMC7196827
- DOI: 10.1073/pnas.1921475117
Dynamic coupling of whole-brain neuronal and neurotransmitter systems
Abstract
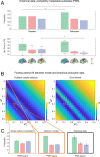
Remarkable progress has come from whole-brain models linking anatomy and function. Paradoxically, it is not clear how a neuronal dynamical system running in the fixed human anatomical connectome can give rise to the rich changes in the functional repertoire associated with human brain function, which is impossible to explain through long-term plasticity. Neuromodulation evolved to allow for such flexibility by dynamically updating the effectivity of the fixed anatomical connectivity. Here, we introduce a theoretical framework modeling the dynamical mutual coupling between the neuronal and neurotransmitter systems. We demonstrate that this framework is crucial to advance our understanding of whole-brain dynamics by bidirectional coupling of the two systems through combining multimodal neuroimaging data (diffusion magnetic resonance imaging [dMRI], functional magnetic resonance imaging [fMRI], and positron electron tomography [PET]) to explain the functional effects of specific serotoninergic receptor (5-HT2AR) stimulation with psilocybin in healthy humans. This advance provides an understanding of why psilocybin is showing considerable promise as a therapeutic intervention for neuropsychiatric disorders including depression, anxiety, and addiction. Overall, these insights demonstrate that the whole-brain mutual coupling between the neuronal and the neurotransmission systems is essential for understanding the remarkable flexibility of human brain function despite having to rely on fixed anatomical connectivity.
Keywords: PET; neurotransmitter; psilocybin; serotonin; whole-brain modeling.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




Comment in
-
Closing the gap between mind and brain with the dynamic connectome.Proc Natl Acad Sci U S A. 2020 May 5;117(18):9677-9678. doi: 10.1073/pnas.2005329117. Epub 2020 Apr 21. Proc Natl Acad Sci U S A. 2020. PMID: 32317384 Free PMC article. No abstract available.
References
-
- Sporns O., Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 23, 162–171 (2013). - PubMed
-
- Deco G., Tononi G., Boly M., Kringelbach M. L., Rethinking segregation and integration: Contributions of whole-brain modelling. Nat. Rev. Neurosci. 16, 430–439 (2015). - PubMed

