A mobile ELF4 delivers circadian temperature information from shoots to roots
- PMID: 32284549
- PMCID: PMC7197390
- DOI: 10.1038/s41477-020-0634-2
A mobile ELF4 delivers circadian temperature information from shoots to roots
Abstract
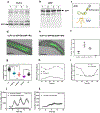
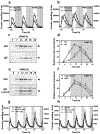
The circadian clock is synchronized by environmental cues, mostly by light and temperature. Explaining how the plant circadian clock responds to temperature oscillations is crucial to understanding plant responsiveness to the environment. Here, we found a prevalent temperature-dependent function of the Arabidopsis clock component EARLY FLOWERING 4 (ELF4) in the root clock. Although the clocks in roots are able to run in the absence of shoots, micrografting assays and mathematical analyses show that ELF4 moves from shoots to regulate rhythms in roots. ELF4 movement does not convey photoperiodic information, but trafficking is essential for controlling the period of the root clock in a temperature-dependent manner. Low temperatures favour ELF4 mobility, resulting in a slow-paced root clock, whereas high temperatures decrease movement, leading to a faster clock. Hence, the mobile ELF4 delivers temperature information and establishes a shoot-to-root dialogue that sets the pace of the clock in roots.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












Comment in
-
ELF in the root.Nat Plants. 2020 Apr;6(4):336-337. doi: 10.1038/s41477-020-0636-0. Nat Plants. 2020. PMID: 32284548 No abstract available.
References
-
- Zhang EE & Kay SA Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell. Biol 11, 764–776 (2010). - PubMed
-
- Greenham K & McClung CR Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet 16, 598–610 (2015). - PubMed
-
- Oakenfull RJ & Davis SJ Shining a light on the Arabidopsis circadian clock. Plant Cell Environ 40, 2571–2585 (2017). - PubMed
-
- Hogenesch JB & Ueda HR Understanding systems-level properties: timely stories from the study of clocks. Nature Reviews Genetics 12, 407 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

