Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury
- PMID: 32284604
- PMCID: PMC7611025
- DOI: 10.1038/s41593-020-0618-6
Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury
Abstract
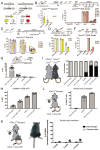
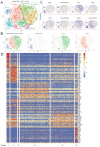
While CNS microglia have been extensively studied, relatively little is known about macrophages populating the peripheral nervous system. Here we performed ontogenic, transcriptomic and spatial characterization of sciatic nerve macrophages (snMacs). Using multiple fate-mapping systems, we show that snMacs do not derive from the early embryonic precursors colonizing the CNS, but originate primarily from late embryonic precursors and become replaced by bone-marrow-derived macrophages over time. Using single-cell transcriptomics, we identified a tissue-specific core signature of snMacs and two spatially separated snMacs: Relmα+Mgl1+ snMacs in the epineurium and Relmα-Mgl1- snMacs in the endoneurium. Globally, snMacs lack most of the core signature genes of microglia, with only the endoneurial subset expressing a restricted number of these genes. In response to nerve injury, the two resident snMac populations respond differently. Moreover, and unlike in the CNS, monocyte-derived macrophages that develop during injury can engraft efficiently in the pool of resident peripheral nervous system macrophages.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

