Regeneration of pulpo-dentinal-like complex by a group of unique multipotent CD24a+ stem cells
- PMID: 32284993
- PMCID: PMC7141825
- DOI: 10.1126/sciadv.aay1514
Regeneration of pulpo-dentinal-like complex by a group of unique multipotent CD24a+ stem cells
Abstract
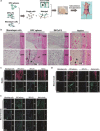
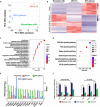
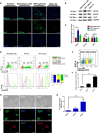
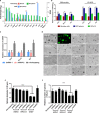
Dental pulp is critical to maintain the vitality of a tooth. Regeneration of pulpo-dentinal complex is of great interest to treat pulpitis and pulp necrosis. In this study, through three-dimensional spheroid culture, a group of unique multipotent stem cells were identified from mouse dental papilla called multipotent dental pulp regenerative stem cells (MDPSCs). MDPSCs exhibited enhanced osteogenic/odontogenic differentiation capabilities and could form regenerative dentin and neurovascular-like structures that mimicked the native teeth in vivo. Further analysis revealed that CD24a was the bona fide marker for MDPSCs, and their expansion was highly dependent on the expression of a key transcriptional factor, Sp7. Last, CD24a+ cells could be detected in primary dental papilla in mice and human, suggesting that MDPSCs resided in their native niches. Together, our study has identified a previously unidentified group of multipotent pulp regenerative stem cells with defined molecular markers for the potential treatment of pulpitis and pulp necrosis.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Simon S. R. J., Berdal A., Cooper P. R., Lumley P. J., Tomson P. L., Smith A. J., Dentin-pulp complex regeneration: From lab to clinic. Adv. Dent. Res. 23, 340–345 (2011). - PubMed
-
- European Society of Endodontology , Quality guidelines for endodontic treatment: consensus report of the European Society of Endodontology. Int. Endod. J. 39, 921–930 (2006). - PubMed
-
- Morsczeck C., Gotz W., Schierholz J., Zeilhofer F., Kuhn U., Mohl C., Sippel C., Hoffmann K. H., Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 24, 155–165 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

