The natverse, a versatile toolbox for combining and analysing neuroanatomical data
- PMID: 32286229
- PMCID: PMC7242028
- DOI: 10.7554/eLife.53350
The natverse, a versatile toolbox for combining and analysing neuroanatomical data
Abstract
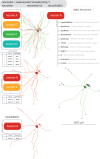
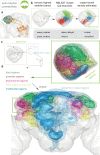
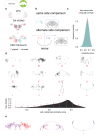
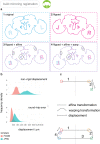
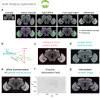
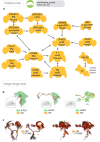
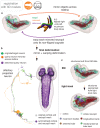
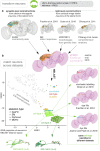
To analyse neuron data at scale, neuroscientists expend substantial effort reading documentation, installing dependencies and moving between analysis and visualisation environments. To facilitate this, we have developed a suite of interoperable open-source R packages called the <monospace>natverse</monospace>. The <monospace>natverse</monospace> allows users to read local and remote data, perform popular analyses including visualisation and clustering and graph-theoretic analysis of neuronal branching. Unlike most tools, the <monospace>natverse</monospace> enables comparison across many neurons of morphology and connectivity after imaging or co-registration within a common template space. The <monospace>natverse</monospace> also enables transformations between different template spaces and imaging modalities. We demonstrate tools that integrate the vast majority of Drosophila neuroanatomical light microscopy and electron microscopy connectomic datasets. The <monospace>natverse</monospace> is an easy-to-use environment for neuroscientists to solve complex, large-scale analysis challenges as well as an open platform to create new code and packages to share with the community.
Keywords: D. melanogaster; analysis software; computational biology; connectomics; mouse; neural circuits; neuroanatomy; neuronal morphology; neuroscience; open-source; systems biology; zebrafish.
© 2020, Bates et al.
Conflict of interest statement
AB, JM, SJ, MC, PS, TR, GJ No competing interests declared
Figures











References
-
- Ascoli GA, Donohue DE, Halavi M. NeuroMorpho.Org: a central resource for neuronal morphologies. Journal of Neuroscience. 2007;27:9247–9251. doi: 10.1523/JNEUROSCI.2055-07.2007. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

