Characterizing Emerging Canine H3 Influenza Viruses
- PMID: 32287326
- PMCID: PMC7182277
- DOI: 10.1371/journal.ppat.1008409
Characterizing Emerging Canine H3 Influenza Viruses
Abstract
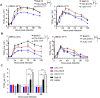
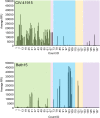
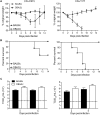
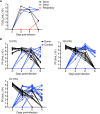
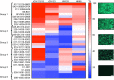
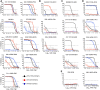
The continual emergence of novel influenza A strains from non-human hosts requires constant vigilance and the need for ongoing research to identify strains that may pose a human public health risk. Since 1999, canine H3 influenza A viruses (CIVs) have caused many thousands or millions of respiratory infections in dogs in the United States. While no human infections with CIVs have been reported to date, these viruses could pose a zoonotic risk. In these studies, the National Institutes of Allergy and Infectious Diseases (NIAID) Centers of Excellence for Influenza Research and Surveillance (CEIRS) network collaboratively demonstrated that CIVs replicated in some primary human cells and transmitted effectively in mammalian models. While people born after 1970 had little or no pre-existing humoral immunity against CIVs, the viruses were sensitive to existing antivirals and we identified a panel of H3 cross-reactive human monoclonal antibodies (hmAbs) that could have prophylactic and/or therapeutic value. Our data predict these CIVs posed a low risk to humans. Importantly, we showed that the CEIRS network could work together to provide basic research information important for characterizing emerging influenza viruses, although there were valuable lessons learned.
Conflict of interest statement
AG-S. is inventor of patents on influenza virus vaccines owned by the Icahn School for Medicine at Mount Sinai and licensed to Medimmune, BI Vetmedica, Vivaldi Biosciences, Zoetis and Avimex.
Figures
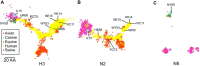




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

