Carnitine palmitoyltransferase 1C contributes to progressive cellular senescence
- PMID: 32289751
- PMCID: PMC7202531
- DOI: 10.18632/aging.103033
Carnitine palmitoyltransferase 1C contributes to progressive cellular senescence
Abstract
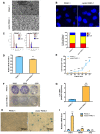
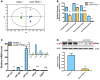
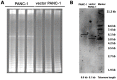
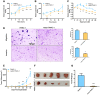
Stable transfection manipulation with antibiotic selection and passaging induces progressive cellular senescence phenotypes. However, the underlying mechanisms remain poorly understood. This study demonstrated that stable transfection of the empty vector induced PANC-1 cells into cellular senescence. Metabolomics revealed several acylcarnitines and their upstream regulatory gene, carnitine palmitoyltransferase 1C (CPT1C) involved in fatty acid β-oxidation in mitochondria, were strikingly decreased in senescent PANC-1 cells. Low CPT1C expression triggered mitochondrial dysfunction, inhibited telomere elongation, impaired cell survival under metabolic stress, and hindered the malignance and tumorigenesis of senescent cells. On the contrary, mitochondrial activity was restored by CPT1C gain-of-function in senescent vector PANC-1 cells. PPARα and TP53/CDKN1A, crucial signaling components in cellular senescence, were downregulated in senescent PANC-1 cells. This study identifies CPT1C as a key regulator of stable transfection-induced progressive PANC-1 cell senescence that inhibits mitochondrial function-associated metabolic reprogramming. These findings confirm the need to identify cell culture alterations after stable transfection, particularly when cells are used for metabolomics and mitochondria-associated studies, and suggest inhibition of CPT1C could be a promising target to intervene pancreatic tumorigenesis.
Keywords: carnitine palmitoyltransferase 1C; metabolic reprogramming; mitochondria; senescence; stable transfection.
Conflict of interest statement
Figures

References
-
- Wang Y, Chen Y, Guan L, Zhang H, Huang Y, Johnson CH, Wu Z, Gonzalez FJ, Yu A, Huang P, Wang Y, Yang S, Chen P, et al.. Carnitine palmitoyltransferase 1C regulates cancer cell senescence through mitochondria-associated metabolic reprograming. Cell Death Differ. 2018; 25:735–48. 10.1038/s41418-017-0013-3 - DOI - PMC - PubMed
-
- Bi HC, Pan YZ, Qiu JX, Krausz KW, Li F, Johnson CH, Jiang CT, Gonzalez FJ, Yu AM. N-methylnicotinamide and nicotinamide N-methyltransferase are associated with microRNA-1291-altered pancreatic carcinoma cell metabolome and suppressed tumorigenesis. Carcinogenesis. 2014; 35:2264–72. 10.1093/carcin/bgu174 - DOI - PMC - PubMed
-
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, Kogan SC, Lowe SW. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011; 25:2125–36. 10.1101/gad.17276711 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

