Global Regulatory Roles of the Histidine-Responsive Transcriptional Repressor HutC in Pseudomonas fluorescens SBW25
- PMID: 32291279
- PMCID: PMC7283599
- DOI: 10.1128/JB.00792-19
Global Regulatory Roles of the Histidine-Responsive Transcriptional Repressor HutC in Pseudomonas fluorescens SBW25
Abstract
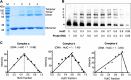
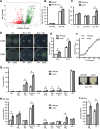
HutC is known as a transcriptional repressor specific for histidine utilization (hut) genes in Gram-negative bacteria, including Pseudomonas fluorescens SBW25. However, its precise mode of protein-DNA interactions hasn't been examined with purified HutC proteins. Here, we performed electrophoretic mobility shift assay (EMSA) and DNase I footprinting using His6-tagged HutC and biotin-labeled probe of the hut promoter (PhutU). Results revealed a complex pattern of HutC oligomerization, and the specific protein-DNA interaction is disrupted by urocanate, a histidine derivative, in a concentration-dependent manner. Next, we searched for putative HutC-binding sites in the SBW25 genome. This led to the identification of 143 candidate targets with a P value less than 10-4 HutC interaction with eight selected candidate sites was subsequently confirmed by EMSA analysis, including the type IV pilus assembly protein PilZ, phospholipase C (PlcC) for phosphatidylcholine hydrolyzation, and key regulators of cellular nitrogen metabolism (NtrBC and GlnE). Finally, an isogenic hutC deletion mutant was subjected to transcriptome sequencing (RNA-seq) analysis and phenotypic characterization. When bacteria were grown on succinate and histidine, hutC deletion caused upregulation of 794 genes and downregulation of 525 genes at a P value of <0.05 with a fold change cutoff of 2.0. The hutC mutant displayed an enhanced spreading motility and pyoverdine production in laboratory media, in addition to the previously reported growth defect on the surfaces of plants. Together, our data indicate that HutC plays global regulatory roles beyond histidine catabolism through low-affinity binding with operator sites located outside the hut locus.IMPORTANCE HutC in Pseudomonas is a representative member of the GntR/HutC family of transcriptional regulators, which possess a N-terminal winged helix-turn-helix (wHTH) DNA-binding domain and a C-terminal substrate-binding domain. HutC is generally known to repress expression of histidine utilization (hut) genes through binding to the PhutU promoter with urocanate (the first intermediate of the histidine degradation pathway) as the direct inducer. Here, we first describe the detailed molecular interactions between HutC and its PhutU target site in a plant growth-promoting bacterium, P. fluorescens SBW25, and further show that HutC possesses specific DNA-binding activities with many targets in the SBW25 genome. Subsequent RNA-seq analysis and phenotypic assays revealed an unexpected global regulatory role of HutC for successful bacterial colonization in planta.
Keywords: HutC; Pseudomonas; gene regulation; histidine utilization; plant-microbe interactions.
Copyright © 2020 American Society for Microbiology.
Figures





References
-
- Balleza E, López-Bojorquez LN, Martínez-Antonio A, Resendis-Antonio O, Lozada-Chávez I, Balderas-Martínez YI, Encarnación S, Collado-Vides J. 2009. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev 33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. - DOI - PMC - PubMed
-
- Itoh Y, Nishijyo T, Nakada Y. 2007. Histidine catabolism and catabolite regulation, p 371–395. In Ramos H-L, Fillous A (ed), Pseudomonas. Springer, Berlin, Germany.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

