Histone methyltransferase SETD1A interacts with HIF1α to enhance glycolysis and promote cancer progression in gastric cancer
- PMID: 32291851
- PMCID: PMC7266269
- DOI: 10.1002/1878-0261.12689
Histone methyltransferase SETD1A interacts with HIF1α to enhance glycolysis and promote cancer progression in gastric cancer
Abstract
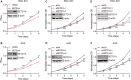
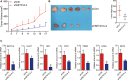
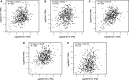
Growing tumors alter their metabolic profiles to support the increased cell proliferation. SETD1A, a histone lysine methyltransferase which specifically methylates H3K4, plays important roles in both normal cell and cancer cell functions. However, the function of SETD1A in gastric cancer (GC) progression and its role in GC metabolic reprogramming are still largely unknown. In the current study, we discovered that the expression of SETD1A was higher in GC tumor specimens compared to surrounding nontumor tissues. Upregulation of SETD1A increased GC cell proliferation, whereas downregulation of SETD1A inhibited GC cell proliferation. Furthermore, knockdown of SETD1A reduced glucose uptake and production of lactate and suppressed glycolysis by decreasing the expression of glycolytic genes, including GLUT1, HK2, PFK2, PKM2, LDHA, and MCT4. Mechanistically, SETD1A interacted with HIF1α to strengthen its transactivation, indicating that SETD1A promotes glycolysis through coactivation of HIF1α. SETD1A and HIF1α were recruited to the promoter of HK2 and PFK2, where SETD1A could methylate H3K4. However, knockdown of SETD1A decreased the methylation of H3K4 on HK2 and PFK2 promoter and reduced HIF1α recruitment necessary to promote transcription of glycolytic genes. Inhibition of HIF1α decelerated SETD1A-enhanced GC cell growth. In additional, there was a linear correlation between SETD1A and several key glycolytic genes in human GC specimens obtained from TCGA dataset. Thus, our results demonstrated that SETD1A interacted with HIF1α to promote glycolysis and accelerate GC progression, implicating that SETD1A may be a potential molecular target for GC treatment.
Keywords: HIF1α; gastric cancer; glycolysis; histone methyltransferase SETD1A; progression.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declared no conflict of interest.
Figures




Similar articles
-
Histone demethylase JMJD1A promotes urinary bladder cancer progression by enhancing glycolysis through coactivation of hypoxia inducible factor 1α.Oncogene. 2017 Jul 6;36(27):3868-3877. doi: 10.1038/onc.2017.13. Epub 2017 Mar 6. Oncogene. 2017. PMID: 28263974
-
Transcription factor LHX9 (LIM Homeobox 9) enhances pyruvate kinase PKM2 activity to induce glycolytic metabolic reprogramming in cancer stem cells, promoting gastric cancer progression.J Transl Med. 2023 Nov 18;21(1):833. doi: 10.1186/s12967-023-04658-7. J Transl Med. 2023. PMID: 37980488 Free PMC article.
-
Non-catalytic role of SETD1A promotes gastric cancer cell proliferation through the E2F4-TAF6 axis in the cell cycle.Cell Death Dis. 2025 Aug 23;16(1):639. doi: 10.1038/s41419-025-07976-4. Cell Death Dis. 2025. PMID: 40846851 Free PMC article.
-
Metabolic phenotype of bladder cancer.Cancer Treat Rev. 2016 Apr;45:46-57. doi: 10.1016/j.ctrv.2016.03.005. Epub 2016 Mar 8. Cancer Treat Rev. 2016. PMID: 26975021 Review.
-
HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms.Mini Rev Med Chem. 2009 Aug;9(9):1084-101. doi: 10.2174/138955709788922610. Mini Rev Med Chem. 2009. PMID: 19689405 Review.
Cited by
-
Transplantation of gastric epithelial mitochondria into human gastric cancer cells inhibits tumor growth and enhances chemosensitivity by reducing cancer stemness and modulating gastric cancer metabolism.Stem Cell Res Ther. 2025 Feb 23;16(1):87. doi: 10.1186/s13287-025-04223-7. Stem Cell Res Ther. 2025. PMID: 39988680 Free PMC article.
-
Roles of Lysine Methylation in Glucose and Lipid Metabolism: Functions, Regulatory Mechanisms, and Therapeutic Implications.Biomolecules. 2024 Jul 19;14(7):862. doi: 10.3390/biom14070862. Biomolecules. 2024. PMID: 39062577 Free PMC article. Review.
-
An SETD1A/Wnt/β-catenin feedback loop promotes NSCLC development.J Exp Clin Cancer Res. 2021 Oct 13;40(1):318. doi: 10.1186/s13046-021-02119-x. J Exp Clin Cancer Res. 2021. PMID: 34645486 Free PMC article.
-
Integration of Epigenetic Mechanisms into Non-Genotoxic Carcinogenicity Hazard Assessment: Focus on DNA Methylation and Histone Modifications.Int J Mol Sci. 2021 Oct 11;22(20):10969. doi: 10.3390/ijms222010969. Int J Mol Sci. 2021. PMID: 34681626 Free PMC article. Review.
-
SCARA5 inhibits gastric cancer progression via epithelial-mesenchymal transition suppression.J Cancer. 2021 Mar 1;12(8):2412-2421. doi: 10.7150/jca.52426. eCollection 2021. J Cancer. 2021. PMID: 33758617 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. - PubMed
-
- Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, Tu Q, Yin D, Lin D, Wong PP et al (2019) Extracellular vesicle‐packaged HIF‐1alpha‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 21, 498–510. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ and He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66, 115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

