ECuADOR-Easy Curation of Angiosperm Duplicated Organellar Regions, a tool for cleaning and curating plastomes assembled from next generation sequencing pipelines
- PMID: 32292644
- PMCID: PMC7147433
- DOI: 10.7717/peerj.8699
ECuADOR-Easy Curation of Angiosperm Duplicated Organellar Regions, a tool for cleaning and curating plastomes assembled from next generation sequencing pipelines
Abstract
Background: With the rapid increase in availability of genomic resources offered by Next-Generation Sequencing (NGS) and the availability of free online genomic databases, efficient and standardized metadata curation approaches have become increasingly critical for the post-processing stages of biological data. Especially in organelle-based studies using circular chloroplast genome datasets, the assembly of the main structural regions in random order and orientation represents a major limitation in our ability to easily generate "ready-to-align" datasets for phylogenetic reconstruction, at both small and large taxonomic scales. In addition, current practices discard the most variable regions of the genomes to facilitate the alignment of the remaining coding regions. Nevertheless, no software is currently available to perform curation to such a degree, through simple detection, organization and positioning of the main plastome regions, making it a time-consuming and error-prone process. Here we introduce a fast and user friendly software ECuADOR, a Perl script specifically designed to automate the detection and reorganization of newly assembled plastomes obtained from any source available (NGS, sanger sequencing or assembler output).
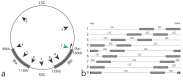
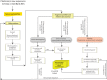
Methods: ECuADOR uses a sliding-window approach to detect long repeated sequences in draft sequences, which then identifies the inverted repeat regions (IRs), even in case of artifactual breaks or sequencing errors and automates the rearrangement of the sequence to the widely used LSC-Irb-SSC-IRa order. This facilitates rapid post-editing steps such as creation of genome alignments, detection of variable regions, SNP detection and phylogenomic analyses.
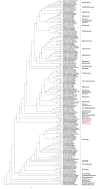
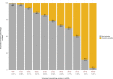
Results: ECuADOR was successfully tested on plant families throughout the angiosperm phylogeny by curating 161 chloroplast datasets. ECuADOR first identified and reordered the central regions (LSC-Irb-SSC-IRa) for each dataset and then produced a new annotation for the chloroplast sequences. The process took less than 20 min with a maximum memory requirement of 150 MB and an accuracy of over 99%.
Conclusions: ECuADOR is the sole de novo one-step recognition and re-ordination tool that provides facilitation in the post-processing analysis of the extra nuclear genomes from NGS data. The program is available at https://github.com/BiodivGenomic/ECuADOR/.
Keywords: Automated workflow; Bioinformatics; NGS; Phylogenomics; Plastome; Sliding window.
© 2020 Armijos Carrion et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Chase MW, Christenhusz MJ, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society. 2016;181(1):1–20. doi: 10.1111/boj.12385. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

