Molecular characterization and expression analysis of pitaya (Hylocereus polyrhizus) HpLRR genes in response to Neoscytalidium dimidiatum infection
- PMID: 32293269
- PMCID: PMC7161156
- DOI: 10.1186/s12870-020-02368-6
Molecular characterization and expression analysis of pitaya (Hylocereus polyrhizus) HpLRR genes in response to Neoscytalidium dimidiatum infection
Abstract
Background: Canker disease caused by Neoscytalidium dimidiatum is a devastating disease resulting in a major loss to the pitaya industry. However, resistance proteins in plants play crucial roles to against pathogen infection. Among resistance proteins, the leucine-rich repeat (LRR) protein is a major family that plays crucial roles in plant growth, development, and biotic and abiotic stress responses, especially in disease defense.
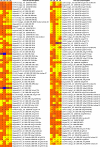
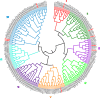
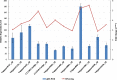
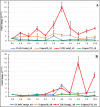
Results: In the present study, a transcriptomics analysis identified a total of 272 LRR genes, 233 of which had coding sequences (CDSs), in the plant pitaya (Hylocereus polyrhizus) in response to fungal Neoscytalidium dimidiatum infection. These genes were divided into various subgroups based on specific domains and phylogenetic analysis. Molecular characterization, functional annotation of proteins, and an expression analysis of the LRR genes were conducted. Additionally, four LRR genes (CL445.Contig4_All, Unigene28_All, CL28.Contig2_All, and Unigene2712_All, which were selected because they had the four longest CDSs were further assessed using quantitative reverse transcription PCR (qRT-PCR) at different fungal infection stages in different pitaya species (Hylocereus polyrhizus and Hylocereus undatus), in different pitaya tissues, and after treatment with salicylic acid (SA), methyl jasmonate (MeJA), and abscisic acid (ABA) hormones. The associated protein functions and roles in signaling pathways were identified.
Conclusions: This study provides a comprehensive overview of the HpLRR family genes at transcriptional level in pitaya in response to N. dimidiatum infection, it will be helpful to understand the molecular mechanism of pitaya canker disease, and lay a strong foundation for further research.
Keywords: Canker disease; Expression analysis; Leucine-rich-repeat genes; Neoscytalidium dimidiatum; Pitaya; Transcriptomics; qRT-PCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- RZZX201904/the Construction of World First Class Discipline of Hainan University
- 18190033-1.Germplasm Protection of Pitaya/Species and Varieties Resources Protection Project of Ministry of Agriculture and Rural Affairs of China
- ZWCX2018007/the Crop Science Postgraduate Innovation Project of Hainan University Tropical Agriculture and Forestry College
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

