New Solutions to Old Problems: Molecular Mechanisms of Meiotic Crossover Control
- PMID: 32294414
- PMCID: PMC7162993
- DOI: 10.1016/j.tig.2020.02.002
New Solutions to Old Problems: Molecular Mechanisms of Meiotic Crossover Control
Abstract
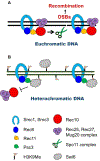
During scientific investigations, the explanation of remarkably interesting phenomena must often await development of new methods or accrual of new observations that in retrospect can lead to lucid answers to the initial problem. A case in point is the control of genetic recombination during meiosis, which leads to crossovers between chromosomes critical for production of healthy offspring. Crossovers must be properly placed along meiotic chromosomes for their accurate segregation. Here, we review observations on two aspects of meiotic crossover control - crossover interference and repression of crossovers near centromeres, both observed more than 85 years ago. Only recently have relatively simple molecular mechanisms for these phenomena become clear through advances in both methods and understanding the molecular basis of meiotic recombination.
Keywords: DNA break hotspot clusters; centromeric repression; crossover interference; heterochromatin; linear element proteins; sister chromatid cohesins.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures

Similar articles
-
Repression of harmful meiotic recombination in centromeric regions.Semin Cell Dev Biol. 2016 Jun;54:188-97. doi: 10.1016/j.semcdb.2016.01.042. Epub 2016 Feb 3. Semin Cell Dev Biol. 2016. PMID: 26849908 Free PMC article. Review.
-
Excess crossovers impede faithful meiotic chromosome segregation in C. elegans.PLoS Genet. 2020 Sep 4;16(9):e1009001. doi: 10.1371/journal.pgen.1009001. eCollection 2020 Sep. PLoS Genet. 2020. PMID: 32886661 Free PMC article.
-
DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis.Genes Dev. 2015 Oct 15;29(20):2183-202. doi: 10.1101/gad.270876.115. Genes Dev. 2015. PMID: 26494791 Free PMC article.
-
Caenorhabditis elegans RMI2 functional homolog-2 (RMIF-2) and RMI1 (RMH-1) have both overlapping and distinct meiotic functions within the BTR complex.PLoS Genet. 2021 Jul 12;17(7):e1009663. doi: 10.1371/journal.pgen.1009663. eCollection 2021 Jul. PLoS Genet. 2021. PMID: 34252074 Free PMC article.
-
The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots?Exp Cell Res. 2012 Jul 15;318(12):1347-52. doi: 10.1016/j.yexcr.2012.03.025. Epub 2012 Mar 31. Exp Cell Res. 2012. PMID: 22487095 Review.
Cited by
-
Exploring chromosomal variations in garden roses: Insights from high-density SNP array data and a new tool, Qploidy.Plant Genome. 2025 Jun;18(2):e70044. doi: 10.1002/tpg2.70044. Plant Genome. 2025. PMID: 40551002 Free PMC article.
-
Genetic Differentiation is Constrained to Chromosomal Inversions and Putative Centromeres in Locally Adapted Populations With Higher Gene Flow.Mol Biol Evol. 2025 Apr 30;42(5):msaf092. doi: 10.1093/molbev/msaf092. Mol Biol Evol. 2025. PMID: 40247662 Free PMC article.
-
Meiotic Crossover Patterning.Front Cell Dev Biol. 2021 Jul 22;9:681123. doi: 10.3389/fcell.2021.681123. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34368131 Free PMC article. Review.
-
Crossover patterns under meiotic chromosome program.Asian J Androl. 2021 Nov-Dec;23(6):562-571. doi: 10.4103/aja.aja_86_20. Asian J Androl. 2021. PMID: 33533735 Free PMC article. Review.
-
From Microscopy to Nanoscopy: Defining an Arabidopsis thaliana Meiotic Atlas at the Nanometer Scale.Front Plant Sci. 2021 May 18;12:672914. doi: 10.3389/fpls.2021.672914. eCollection 2021. Front Plant Sci. 2021. PMID: 34084178 Free PMC article. Review.
References
-
- Jang JK, et al. (1995) Induction of metaphase arrest in Drosophila oocytes by chiasma-based kinetochore tension. Science 268, 1917–1919 - PubMed
-
- Li X and Nicklas RB (1995) Mitotic forces control a cell-cycle checkpoint. Nature 373, 630–632 - PubMed
-
- Koehler KE, et al. (1996) Recombination and nondisjunction in humans and flies. Human Molecular Genetics 5, 1495–1504 - PubMed
-
- Hassold T and Chiu D (1985) Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 70, 11–17 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

