Molecular Investigation of SARS-CoV-2 Proteins and Their Interactions with Antiviral Drugs
- PMID: 32295237
- PMCID: PMC7232184
- DOI: 10.3390/v12040445
Molecular Investigation of SARS-CoV-2 Proteins and Their Interactions with Antiviral Drugs
Abstract
A new Coronavirus strain, named SARS-CoV-2, suddenly emerged in early December 2019. SARS-CoV-2 resulted in being dramatically infectious, with thousands of people infected. In this scenario, and without effective vaccines available, the importance of an immediate tool to support patients and against viral diffusion becomes evident. In this study, we exploit the molecular docking approach to analyze the affinity between different viral proteins and several inhibitors, originally developed for other viral infections. Our data show that, in some cases, a relevant binding can be detected. These findings support the hypothesis to develop new antiviral agents against COVID-19, on the basis of already established therapies.
Keywords: COVID-19; RNA-dependent RNA-polymerase; SARS–CoV-2; antiviral drug; coronavirus; molecular docking; molecular modeling; spike protein; viral protease; viral protein N.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
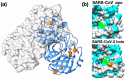
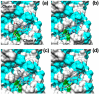
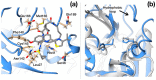


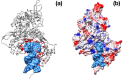

References
-
- Höfer C.T., Di Lella S., Dahmani I., Jungnick N., Bordag N., Bobone S., Huang Q., Keller S., Herrmann A., Chiantia S. Structural determinants of the interaction between influenza A virus matrix protein M1 and lipid membranes. Biochim. Biophys. Acta Biomembr. 2019;1861:1123–1134. doi: 10.1016/j.bbamem.2019.03.013. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

