Identification and Characterization of microRNAs in the Developing Seed of Linseed Flax (Linum usitatissimum L.)
- PMID: 32295287
- PMCID: PMC7215410
- DOI: 10.3390/ijms21082708
Identification and Characterization of microRNAs in the Developing Seed of Linseed Flax (Linum usitatissimum L.)
Abstract
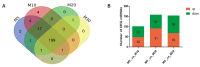
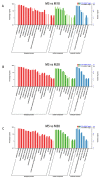
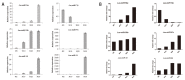
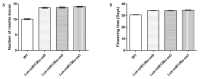
Seed development plays an important role during the life cycle of plants. Linseed flax is an oil crop and the seed is a key organ for fatty acids synthesis and storage. So it is important to understand the molecular mechanism of fatty acid biosynthesis during seed development. In this study, four small RNA libraries from early seeds at 5, 10, 20 and 30 days after flowering (DAF) were constructed and used for high-throughput sequencing to identify microRNAs (miRNAs). A total of 235 miRNAs including 114 known conserved miRNAs and 121 novel miRNAs were identified. The expression patterns of these miRNAs in the four libraries were investigated by bioinformatics and quantitative real-time polymerase chain reaction (qPCR) analysis. It was found that several miRNAs, including Lus-miRNA156a was significantly correlated with seed development process. In order to confirm the actual biological function of Lus-miRNA156a, over-expression vector was constructed and transformed to Arabidopsis. The phenotypes of homozygous transgenic lines showed decreasing of oil content and most of the fatty acid content in seeds as well as late flowering time. The results provided a clue that miRNA156a participating the fatty acid biosynthesis pathway and the detailed molecular mechanism of how it regulates the pathway needs to be further investigated.
Keywords: fatty acid synthesis; linseed flax; miRNA156; microRNA; seed development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

