Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study
- PMID: 32295479
- PMCID: PMC7196925
- DOI: 10.1080/07391102.2020.1756411
Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study
Abstract
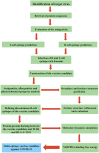
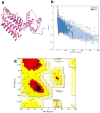
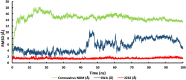
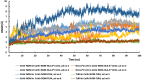
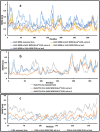
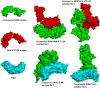
At present, novel Coronavirus (2019-nCoV, the causative agent of COVID-19) has caused worldwide social and economic disruption. The disturbing statistics of this infection promoted us to develop an effective vaccine candidate against the COVID-19. In this study, bioinformatics approaches were employed to design and introduce a novel multi-epitope vaccine against 2019-nCoV that can potentially trigger both CD4+ and CD8+ T-cell immune responses and investigated its biological activities by computational tools. Three known antigenic proteins (Nucleocapsid, ORF3a, and Membrane protein, hereafter called NOM) from the virus were selected and analyzed for prediction of the potential immunogenic B and T-cell epitopes and then validated using bioinformatics tools. Based on in silico analysis, we have constructed a multi-epitope vaccine candidate (NOM) with five rich-epitopes domain including highly scored T and B-cell epitopes. After predicting and evaluating of the third structure of the protein candidate, the best 3 D predicted model was applied for docking studies with Toll-like receptor 4 (TLR4) and HLA-A*11:01. In the next step, molecular dynamics (MD) simulation was used to evaluate the stability of the designed fusion protein with TLR4 and HLA-A*11:01 receptors. MD studies demonstrated that the NOM-TLR4 and NOM-HLA-A*11:01 docked models were stable during simulation time. In silico evaluation showed that the designed chimeric protein could simultaneously elicit humoral and cell-mediated immune responses. Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Coronavirus; Epitope; Immunoinformatics; Vaccine.
Figures



References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., & Lindahl E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
