Omics Data Reveal Putative Regulators of Einkorn Grain Protein Composition under Sulfur Deficiency
- PMID: 32295821
- PMCID: PMC7271774
- DOI: 10.1104/pp.19.00842
Omics Data Reveal Putative Regulators of Einkorn Grain Protein Composition under Sulfur Deficiency
Abstract
Understanding the molecular mechanisms controlling the accumulation of grain storage proteins in response to nitrogen (N) and sulfur (S) nutrition is essential to improve cereal grain nutritional and functional properties. Here, we studied the grain transcriptome and metabolome responses to postanthesis N and S supply for the diploid wheat einkorn (Triticum monococcum). During grain filling, 848 transcripts and 24 metabolites were differentially accumulated in response to N and S availability. The accumulation of total free amino acids per grain and the expression levels of 241 genes showed significant modifications during most of the grain filling period and were upregulated in response to S deficiency. Among them, 24 transcripts strongly responded to S deficiency and were identified in coexpression network analyses as potential coordinators of the grain response to N and S supply. Sulfate transporters and genes involved in sulfate and Met metabolism were upregulated, suggesting regulation of the pool of free amino acids and of the grain N-to-S ratio. Several genes highlighted in this study might limit the impact of S deficiency on the accumulation of grain storage proteins.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures

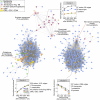
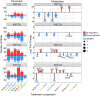



References
-
- Aguirrezábal L, Martre P, Pereyra-Irujo G, Echarte MM, Izquierdo N (2015) Improving grain quality: Ecophysiological and modeling tools to develop management and breeding strategies. In Sadras VO, Calderini DF, eds, Crop Physiology: Applications for Genetic Improvement and Agronomy, Ed 2. Academic Press, Amsterdam, pp 423–465
-
- Allwood JW, De Vos RCH, Moing A, Deborde C, Erban A, Kopka J, Goodacre R, Hall RD (2011) Plant metabolomics and its potential for systems biology research: Background concepts, technology, and methodology. In Jameson D, Verma M, Westerhoff HV, eds, Methods in Systems Biology, Methods in Enzymology, Vol 500. Elsevier, Amsterdam, pp 299–336 - PubMed
-
- Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J, et al. ; International Wheat Genome Sequencing Consortium I (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. - PubMed
-
- Banerjee A, Roychoudhury A(2019) Role of glutathione in plant abiotic stress tolerance In Khan NA, Singh S, and Umar S, eds, Sulfur Assimilation and Abiotic Stress in Plants. Springer, Berlin/Heidelberg, pp 207–225
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

