A long lost key opens an ancient lock: Drosophila Myb causes a synthetic multivulval phenotype in nematodes
- PMID: 32295830
- PMCID: PMC7225089
- DOI: 10.1242/bio.051508
A long lost key opens an ancient lock: Drosophila Myb causes a synthetic multivulval phenotype in nematodes
Abstract
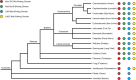
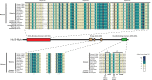
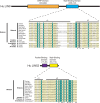
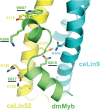
The five-protein MuvB core complex is highly conserved in animals. This nuclear complex interacts with RB-family tumor suppressor proteins and E2F-DP transcription factors to form DREAM complexes that repress genes that regulate cell cycle progression and cell fate. The MuvB core complex also interacts with Myb family oncoproteins to form the Myb-MuvB complexes that activate many of the same genes. We show that animal-type Myb genes are present in Bilateria, Cnidaria and Placozoa, the latter including the simplest known animal species. However, bilaterian nematode worms lost their animal-type Myb genes hundreds of millions of years ago. Nevertheless, amino acids in the LIN9 and LIN52 proteins that directly interact with the MuvB-binding domains of human B-Myb and Drosophila Myb are conserved in Caenorhabditiselegans Here, we show that, despite greater than 500 million years since their last common ancestor, the Drosophila melanogaster Myb protein can bind to the nematode LIN9-LIN52 proteins in vitro and can cause a synthetic multivulval (synMuv) phenotype in vivo This phenotype is similar to that caused by loss-of-function mutations in C. elegans synMuvB-class genes including those that encode homologs of the MuvB core, RB, E2F and DP. Furthermore, amino acid substitutions in the MuvB-binding domain of Drosophila Myb that disrupt its functions in vitro and in vivo also disrupt these activities in C. elegans We speculate that nematodes and other animals may contain another protein that can bind to LIN9 and LIN52 in order to activate transcription of genes repressed by DREAM complexes.
Keywords: Development; Evolution; Myb; Oncogene; Tumor suppressor; synMuv.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Amatschek S., Koenig U., Auer H., Steinlein P., Pacher M., Gruenfelder A., Dekan G., Vogl S., Kubista E., Heider K.-H. et al. (2004). Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 64, 844-856. 10.1158/0008-5472.CAN-03-2361 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

