IACS-010759, a potent inhibitor of glycolysis-deficient hypoxic tumor cells, inhibits mitochondrial respiratory complex I through a unique mechanism
- PMID: 32295842
- PMCID: PMC7247293
- DOI: 10.1074/jbc.RA120.013366
IACS-010759, a potent inhibitor of glycolysis-deficient hypoxic tumor cells, inhibits mitochondrial respiratory complex I through a unique mechanism
Abstract
The small molecule IACS-010759 has been reported to potently inhibit the proliferation of glycolysis-deficient hypoxic tumor cells by interfering with the functions of mitochondrial NADH-ubiquinone oxidoreductase (complex I) without exhibiting cytotoxicity at tolerated doses in normal cells. Considering the significant cytotoxicity of conventional quinone-site inhibitors of complex I, such as piericidin and acetogenin families, we hypothesized that the mechanism of action of IACS-010759 on complex I differs from that of other known quinone-site inhibitors. To test this possibility, here we investigated IACS-010759's mechanism in bovine heart submitochondrial particles. We found that IACS-010759, like known quinone-site inhibitors, suppresses chemical modification by the tosyl reagent AL1 of Asp160 in the 49-kDa subunit, located deep in the interior of a previously proposed quinone-access channel. However, contrary to the other inhibitors, IACS-010759 direction-dependently inhibited forward and reverse electron transfer and did not suppress binding of the quinazoline-type inhibitor [125I]AzQ to the N terminus of the 49-kDa subunit. Photoaffinity labeling experiments revealed that the photoreactive derivative [125I]IACS-010759-PD1 binds to the middle of the membrane subunit ND1 and that inhibitors that bind to the 49-kDa or PSST subunit cannot suppress the binding. We conclude that IACS-010759's binding location in complex I differs from that of any other known inhibitor of the enzyme. Our findings, along with those from previous study, reveal that the mechanisms of action of complex I inhibitors with widely different chemical properties are more diverse than can be accounted for by the quinone-access channel model proposed by structural biology studies.
Keywords: IACS-010759; bioenergetics; cancer; chemical biology; complex I; enzyme inhibitor; hypoxia; mitochondria; photoaffinity labeling; ubiquinone.
© 2020 Tsuji et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

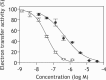
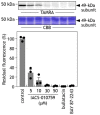
 CH via click chemistry after solubilizing SMPs. Bullatacin (10 μ
CH via click chemistry after solubilizing SMPs. Bullatacin (10 μ
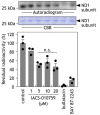

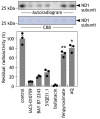

References
-
- Cantor J. R., and Sabatini D. M.. 2012) Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898 10.1158/2159-8290.CD-12-0345 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

