Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants
- PMID: 32296570
- PMCID: PMC7156227
- DOI: 10.1016/j.jpha.2020.03.009
Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants
Abstract
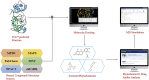
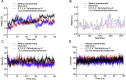
The recent pandemic of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 has raised global health concerns. The viral 3-chymotrypsin-like cysteine protease (3CLpro) enzyme controls coronavirus replication and is essential for its life cycle. 3CLpro is a proven drug discovery target in the case of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Recent studies revealed that the genome sequence of SARS-CoV-2 is very similar to that of SARS-CoV. Therefore, herein, we analysed the 3CLpro sequence, constructed its 3D homology model, and screened it against a medicinal plant library containing 32,297 potential anti-viral phytochemicals/traditional Chinese medicinal compounds. Our analyses revealed that the top nine hits might serve as potential anti- SARS-CoV-2 lead molecules for further optimisation and drug development process to combat COVID-19.
Keywords: COVID-19; Coronavirus; Molecular docking; Molecular dynamics simulation; Natural products; Protein homology modelling; SARS-CoV-2.
© 2020 Xi'an Jiaotong University. Production and hosting by Elsevier B.V.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

