Chaperome Networks - Redundancy and Implications for Cancer Treatment
- PMID: 32297213
- PMCID: PMC7279512
- DOI: 10.1007/978-3-030-40204-4_6
Chaperome Networks - Redundancy and Implications for Cancer Treatment
Abstract
The chaperome is a large family of proteins composed of chaperones, co-chaperones and a multitude of other factors. Elegant studies in yeast and other organisms have paved the road to how we currently understand the complex organization of this large family into protein networks. The goal of this chapter is to provide an overview of chaperome networks in cancer cells, with a focus on two cellular states defined by chaperome network organization. One state characterized by chaperome networks working in isolation and with little overlap, contains global chaperome networks resembling those of normal, non-transformed, cells. We propose that in this state, redundancy in chaperome networks results in a tumor type unamenable for single-agent chaperome therapy. The second state comprises chaperome networks interconnected in response to cellular stress, such as MYC hyperactivation. This is a state where no redundant pathways can be deployed, and is a state of vulnerability, amenable for chaperome therapy. We conclude by proposing a change in how we discover and implement chaperome inhibitor strategies, and suggest an approach to chaperome therapy where the properties of chaperome networks, rather than genetics or client proteins, are used in chaperome inhibitor implementation.
Keywords: Anti-cancer therapy; Chaperome networks; Epichaperome; HSP90 inhibitors; PU-H71; Protein network connectivity; Protein network vulnerability.
Conflict of interest statement
Figures
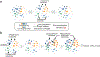

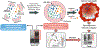
References
-
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124:75–88 - PubMed
-
- Barrios-Rodiles M, Ellis JD, Blencowe BJ, Wrana JL (2017) LUMIER: a discovery tool for mammalian protein interaction networks. Methods Mol Biol 1550:137–148 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

