PU.1 regulates Ccr7 gene expression by binding to its promoter in naïve CD4+ T cells
- PMID: 32297481
- PMCID: PMC7262917
- DOI: 10.1002/2211-5463.12861
PU.1 regulates Ccr7 gene expression by binding to its promoter in naïve CD4+ T cells
Abstract
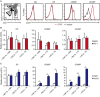
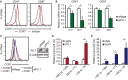
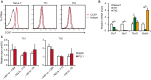
C-C chemokine receptor type 7 (CCR7) is expressed on naïve T cells, B cells, and activated dendritic cells (DCs). We previously demonstrated that the transcription factor PU.1/Spi1 positively regulates the expression of CCR7 in DCs. In the present study, we investigated the role of PU.1 in CCR7 expression in T cells. To confirm whether PU.1 is involved in the expression of CCR7, we conducted a ChIP assay in various T cells purified from splenocytes and thymocytes and found that PU.1 binds to the Ccr7 promoter-proximal region in spleen naïve CD4+ T cells, but not in thymocytes. Small interfering RNA-mediated PU.1 knockdown resulted in decreased CCR7 expression in spleen naïve CD4+ T cells. Compared to naïve CD4+ T cells, Spi1 and Ccr7 mRNA levels decreased in Th1 and Th2 cells, in which PU.1 did not bind to the Ccr7 promoter, suggesting that CCR7 expression decreases due to the dissociation of PU.1 from the Ccr7 promoter during the development of effector T cells from naïve T cells. Collectively, we concluded that CCR7 expression level correlates with the binding level of PU.1 to the Ccr7 promoter and PU.1 acts as a transcriptional activator of the Ccr7 gene in naïve CD4+ T cells.
Keywords: CCR7; Naïve CD4 T cells; PU.1.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Forster R, Davalos‐Misslitz AC and Rot A (2008) CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol 8, 362–371. - PubMed
-
- Nitta T, Murata S, Ueno T, Tanaka K and Takahama Y (2008) Thymic microenvironments for T‐cell repertoire formation. Adv Immunol 99, 59–94. - PubMed
-
- von Andrian UH and Mempel TR (2003) Homing and cellular traffic in lymph nodes. Nat Rev Immunol 3, 867–878. - PubMed
-
- Takada K and Jameson SC (2009) Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol 9, 823–832. - PubMed
-
- Randolph GJ, Angeli V and Swartz MA (2005) Dendritic‐cell trafficking to lymph nodes through lymphatic vessels. Nat Rev Immunol 5, 617–628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

