Microfluidic Platform for the Isolation of Cancer-Cell Subpopulations Based on Single-Cell Glycolysis
- PMID: 32297730
- PMCID: PMC7473507
- DOI: 10.1021/acs.analchem.9b05738
Microfluidic Platform for the Isolation of Cancer-Cell Subpopulations Based on Single-Cell Glycolysis
Abstract
High rates of glycolysis in tumors have been associated with cancer metastasis, tumor recurrence, and poor outcomes. In this light, single cells that exhibit high glycolysis are specific targets for therapy. However, the study of these cells requires efficient tools for their isolation. We use a droplet microfluidic technique developed in our lab,
Figures

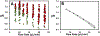

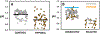
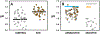
References
-
- Bonnet D; Dick JE Human Acute Myeloid Leukemia Is Organized as a Hierarchy That Originates from a Primitive Hematopoietic Cell. Nat. Med 1997, 3 (7), 730–737. - PubMed
-
- Danhier P; Bański P; Payen VL; Grasso D; Ippolito L; Sonveaux P; Porporato PE Cancer Metabolism in Space and Time: Beyond the Warburg Effect. Biochim. Biophys. Acta - Bioenerg 2017, 1858 (8), 556–572. - PubMed
-
- Herzenberg LA; Parks D; Sahaf B; Perez O; Roederer M; Herzenberg LA The History and Future of the Fluorescence Activated Cell Sorter and Flow Cytometry: A View from Stanford. Clin. Chem, 2002, 48, 1819–1827. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

