Deconstructing cerebellar development cell by cell
- PMID: 32298260
- PMCID: PMC7161948
- DOI: 10.1371/journal.pgen.1008630
Deconstructing cerebellar development cell by cell
Abstract
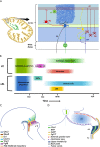
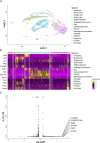
The cerebellum is a pivotal centre for the integration and processing of motor and sensory information. Its extended development into the postnatal period makes this structure vulnerable to a variety of pathologies, including neoplasia. These properties have prompted intensive investigations that reveal not only developmental mechanisms in common with other regions of the neuraxis but also unique strategies to generate neuronal diversity. How the phenotypically distinct cell types of the cerebellum emerge rests on understanding how gene expression differences arise in a spatially and temporally coordinated manner from initially homogeneous cell populations. Increasingly sophisticated fate mapping approaches, culminating in genetic-induced fate mapping, have furthered the understanding of lineage relationships between early- versus later-born cells. Tracing the developmental histories of cells in this way coupled with analysis of gene expression patterns has provided insight into the developmental genetic programmes that instruct cellular heterogeneity. A limitation to date has been the bulk analysis of cells, which blurs lineage relationships and obscures gene expression differences between cells that underpin the cellular taxonomy of the cerebellum. This review emphasises recent discoveries, focusing mainly on single-cell sequencing in mouse and parallel human studies that elucidate neural progenitor developmental trajectories with unprecedented resolution. Complementary functional studies of neural repair after cerebellar injury are challenging assumptions about the stability of postnatal cellular identities. The result is a wealth of new information about the developmental mechanisms that generate cerebellar neural diversity, with implications for human evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

