Pilot Study of Return of Genetic Results to Patients in Adult Nephrology
- PMID: 32299846
- PMCID: PMC7269209
- DOI: 10.2215/CJN.12481019
Pilot Study of Return of Genetic Results to Patients in Adult Nephrology
Abstract
Background and objectives: Actionable genetic findings have implications for care of patients with kidney disease, and genetic testing is an emerging tool in nephrology practice. However, there are scarce data regarding best practices for return of results and clinical application of actionable genetic findings for kidney patients.
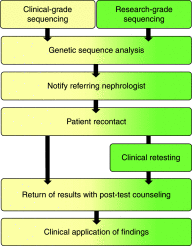
Design, setting, participants, & measurements: We developed a return of results workflow in collaborations with clinicians for the retrospective recontact of adult nephrology patients who had been recruited into a biobank research study for exome sequencing and were identified to have medically actionable genetic findings.
Results: Using this workflow, we attempted to recontact a diverse pilot cohort of 104 nephrology research participants with actionable genetic findings, encompassing 34 different monogenic etiologies of nephropathy and five single-gene disorders recommended by the American College of Medical Genetics and Genomics for return as medically actionable secondary findings. We successfully recontacted 64 (62%) participants and returned results to 41 (39%) individuals. In each case, the genetic diagnosis had meaningful implications for the patients' nephrology care. Through implementation efforts and qualitative interviews with providers, we identified over 20 key challenges associated with returning results to study participants, and found that physician knowledge gaps in genomics was a recurrent theme. We iteratively addressed these challenges to yield an optimized workflow, which included standardized consultation notes with tailored management recommendations, monthly educational conferences on core topics in genomics, and a curated list of expert clinicians for patients requiring extranephrologic referrals.
Conclusions: Developing the infrastructure to support return of genetic results in nephrology was resource-intensive, but presented potential opportunities for improving patient care.
Podcast: This article contains a podcast at https://www.asn-online.org/media/podcast/CJASN/2020_04_16_12481019.mp3.
Keywords: adult; biological specimen banks; chronic kidney disease; cohort studies; exome; familial nephropathy; genetic renal disease; genetic testing; genomics; human genetics; humans; kidney diseases; medical genetics; nephrology; patient care; pilot projects; referral and consultation; retrospective studies; whole exome sequencing; workflow.
Copyright © 2020 by the American Society of Nephrology.
Figures

References
-
- Mallett AJ, McCarthy HJ, Ho G, Holman K, Farnsworth E, Patel C, Fletcher JT, Mallawaarachchi A, Quinlan C, Bennetts B, Alexander SI: Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int 92: 1493–1506, 2017 - PubMed
-
- Warejko JK, Tan W, Daga A, Schapiro D, Lawson JA, Shril S, Lovric S, Ashraf S, Rao J, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, Schneider R, Gee HY, Schmidt JM, Vivante A, van der Ven AT, Ityel H, Chen J, Sadowski CE, Kohl S, Pabst WL, Nakayama M, Somers MJG, Rodig NM, Daouk G, Baum M, Stein DR, Ferguson MA, Traum AZ, Soliman NA, Kari JA, El Desoky S, Fathy H, Zenker M, Bakkaloglu SA, Müller D, Noyan A, Ozaltin F, Cadnapaphornchai MA, Hashmi S, Hopcian J, Kopp JB, Benador N, Bockenhauer D, Bogdanovic R, Stajić N, Chernin G, Ettenger R, Fehrenbach H, Kemper M, Munarriz RL, Podracka L, Büscher R, Serdaroglu E, Tasic V, Mane S, Lifton RP, Braun DA, Hildebrandt F: Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 13: 53–62, 2018 - PMC - PubMed
-
- van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, Vivante A, Hwang DY, Schulz J, Braun DA, Schmidt JM, Schapiro D, Schneider R, Warejko JK, Daga A, Majmundar AJ, Tan W, Jobst-Schwan T, Hermle T, Widmeier E, Ashraf S, Amar A, Hoogstraaten CA, Hugo H, Kitzler TM, Kause F, Kolvenbach CM, Dai R, Spaneas L, Amann K, Stein DR, Baum MA, Somers MJG, Rodig NM, Ferguson MA, Traum AZ, Daouk GH, Bogdanović R, Stajić N, Soliman NA, Kari JA, El Desoky S, Fathy HM, Milosevic D, Al-Saffar M, Awad HS, Eid LA, Selvin A, Senguttuvan P, Sanna-Cherchi S, Rehm HL, MacArthur DG, Lek M, Laricchia KM, Wilson MW, Mane SM, Lifton RP, Lee RS, Bauer SB, Lu W, Reutter HM, Tasic V, Shril S, Hildebrandt F: Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 29: 2348–2361, 2018 - PMC - PubMed
-
- Lata S, Marasa M, Li Y, Fasel DA, Groopman E, Jobanputra V, Rasouly H, Mitrotti A, Westland R, Verbitsky M, Nestor J, Slater LM, D’Agati V, Zaniew M, Materna-Kiryluk A, Lugani F, Caridi G, Rampoldi L, Mattoo A, Newton CA, Rao MK, Radhakrishnan J, Ahn W, Canetta PA, Bomback AS, Appel GB, Antignac C, Markowitz GS, Garcia CK, Kiryluk K, Sanna-Cherchi S, Gharavi AG: Whole-exome sequencing in adults with chronic kidney disease: A pilot study. Ann Intern Med 168: 100–109, 2018 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

