A viral toolkit for recording transcription factor-DNA interactions in live mouse tissues
- PMID: 32300008
- PMCID: PMC7211997
- DOI: 10.1073/pnas.1918241117
A viral toolkit for recording transcription factor-DNA interactions in live mouse tissues
Abstract
Transcription factors (TFs) enact precise regulation of gene expression through site-specific, genome-wide binding. Common methods for TF-occupancy profiling, such as chromatin immunoprecipitation, are limited by requirement of TF-specific antibodies and provide only end-point snapshots of TF binding. Alternatively, TF-tagging techniques, in which a TF is fused to a DNA-modifying enzyme that marks TF-binding events across the genome as they occur, do not require TF-specific antibodies and offer the potential for unique applications, such as recording of TF occupancy over time and cell type specificity through conditional expression of the TF-enzyme fusion. Here, we create a viral toolkit for one such method, calling cards, and demonstrate that these reagents can be delivered to the live mouse brain and used to report TF occupancy. Further, we establish a Cre-dependent calling cards system and, in proof-of-principle experiments, show utility in defining cell type-specific TF profiles and recording and integrating TF-binding events across time. This versatile approach will enable unique studies of TF-mediated gene regulation in live animal models.
Keywords: brain; enhancer; epigenetics; recording; transcription factor.
Conflict of interest statement
Competing interest statement: R.D.M., A.M., and M.N.W. have filed a patent application on SRT technology. No other authors have disclosures to report.
Figures


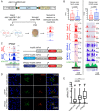

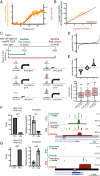
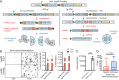
References
-
- Spitz F., Furlong E. E. M., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 NS105363/NS/NINDS NIH HHS/United States
- RF1 MH117070/MH/NIMH NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R21 HG009750/HG/NHGRI NIH HHS/United States
- T32 GM007200/GM/NIGMS NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- R25 GM103757/GM/NIGMS NIH HHS/United States
- T32 GM008151/GM/NIGMS NIH HHS/United States
- F30 HG009986/HG/NHGRI NIH HHS/United States
- T32 HG000045/HG/NHGRI NIH HHS/United States
- T32 GM007067/GM/NIGMS NIH HHS/United States
- U54 HD087011/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

