Transaminases for industrial biocatalysis: novel enzyme discovery
- PMID: 32300853
- PMCID: PMC7228992
- DOI: 10.1007/s00253-020-10585-0
Transaminases for industrial biocatalysis: novel enzyme discovery
Abstract
Transaminases (TAms) are important enzymes for the production of chiral amines for the pharmaceutical and fine chemical industries. Novel TAms for use in these industries have been discovered using a range of approaches, including activity-guided methods and homologous sequence searches from cultured microorganisms to searches using key motifs and metagenomic mining of environmental DNA libraries. This mini-review focuses on the methods used for TAm discovery over the past two decades, analyzing the changing trends in the field and highlighting the advantages and drawbacks of the respective approaches used. This review will also discuss the role of protein engineering in the development of novel TAms and explore possible directions for future TAm discovery for application in industrial biocatalysis. KEY POINTS: • The past two decades of TAm enzyme discovery approaches are explored. • TAm sequences are phylogenetically analyzed and compared to other discovery methods. • Benefits and drawbacks of discovery approaches for novel biocatalysts are discussed. • The role of protein engineering and future discovery directions is highlighted.
Keywords: Biocatalysis; Chiral amine; Enzyme discovery; Metagenomics; Transaminase.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
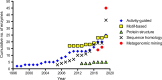
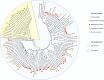

References
-
- Baud D, Jeffries JWE, Moody TS, Ward JM, Hailes HC. A metagenomics approach for new biocatalyst discovery: application to transaminases and the synthesis of allylic amines. Green Chem. 2017;19:1134–1143. doi: 10.1039/c6gc02769e. - DOI
-
- Braun-Galleani S, Henríquez M-J, Nesbeth DN (2019) Whole cell biosynthesis of 1-methyl-3-phenylpropylamine and 2-amino-1,3,4-butanetriol using Komagataella phaffii (Pichia pastoris) strain BG-10 engineered with a transgene encoding Chromobacterium violaceum ω-transaminase. Heliyon 5:e02338. 10.1016/j.heliyon.2019.e02338 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

