The Epithelial adhesin 1 tandem repeat region mediates protein display through multiple mechanisms
- PMID: 32301985
- PMCID: PMC7199969
- DOI: 10.1093/femsyr/foaa018
The Epithelial adhesin 1 tandem repeat region mediates protein display through multiple mechanisms
Abstract
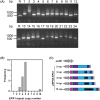
The pathogenic yeast Candida glabrata is reliant on a suite of cell surface adhesins that play a variety of roles necessary for transmission, establishment and proliferation during infection. One particular adhesin, Epithelial Adhesin 1 [Epa1p], is responsible for binding to host tissue, a process which is essential for fungal propagation. Epa1p structure consists of three domains: an N-terminal intercellular binding domain responsible for epithelial cell binding, a C-terminal GPI anchor for cell wall linkage and a serine/threonine-rich linker domain connecting these terminal domains. The linker domain contains a 40-amino acid tandem repeat region, which we have found to be variable in repeat copy number between isolates from clinical sources. We hypothesized that natural variation in Epa1p repeat copy may modulate protein function. To test this, we recombinantly expressed Epa1p with various repeat copy numbers in S. cerevisiae to determine how differences in repeat copy number affect Epa1p expression, surface display and binding to human epithelial cells. Our data suggest that repeat copy number variation has pleiotropic effects, influencing gene expression, protein surface display and shedding from the cell surface of the Epa1p adhesin. This study serves to demonstrate repeat copy number variation can modulate protein function through a number of mechanisms in order to contribute to pathogenicity of C. glabrata.
Keywords: antimicrobial; killer yeasts; mycocins.
© FEMS 2020.
Figures




References
-
- Babokhov M, Reinfeld BI, Hackbarth Ket al.. Tandem repeats drive variation of intrinsically disordered regions in budding yeast. bioRxiv. 2018b;339663.
-
- Biscotti MA, Canapa A, Forconi Met al.. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res. 2015;23:463–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

