G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules
- PMID: 32302571
- PMCID: PMC7448383
- DOI: 10.1016/j.cell.2020.03.046
G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules
Abstract
The mechanisms underlying ribonucleoprotein (RNP) granule assembly, including the basis for establishing and maintaining RNP granules with distinct composition, are unknown. One prominent type of RNP granule is the stress granule (SG), a dynamic and reversible cytoplasmic assembly formed in eukaryotic cells in response to stress. Here, we show that SGs assemble through liquid-liquid phase separation (LLPS) arising from interactions distributed unevenly across a core protein-RNA interaction network. The central node of this network is G3BP1, which functions as a molecular switch that triggers RNA-dependent LLPS in response to a rise in intracellular free RNA concentrations. Moreover, we show that interplay between three distinct intrinsically disordered regions (IDRs) in G3BP1 regulates its intrinsic propensity for LLPS, and this is fine-tuned by phosphorylation within the IDRs. Further regulation of SG assembly arises through positive or negative cooperativity by extrinsic G3BP1-binding factors that strengthen or weaken, respectively, the core SG network.
Keywords: G3BP1; Intrinsically disordered protein; biomolecular condensate; cooperativity; core stress granule network; liquid-liquid phase separation; membraneless organelle; molecular switch; multivalency; stress granule.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.P.T. is a consultant for 5AM and Third Rock Ventures.
Figures
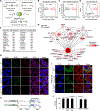

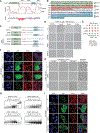
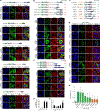

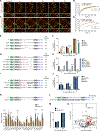

Comment in
-
Networking and Dynamic Switches in Biological Condensates.Cell. 2020 Apr 16;181(2):228-230. doi: 10.1016/j.cell.2020.03.056. Cell. 2020. PMID: 32302565
References
-
- Barabasi AL, and Albert R (1999). Emergence of scaling in random networks. Science 286, 509–512. - PubMed
-
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, and Svergun DI (2007). Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc 129, 5656–5664. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

