DNA polymerase theta (Polθ) - an error-prone polymerase necessary for genome stability
- PMID: 32302896
- PMCID: PMC7230004
- DOI: 10.1016/j.gde.2020.02.017
DNA polymerase theta (Polθ) - an error-prone polymerase necessary for genome stability
Abstract
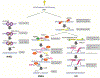
Mammalian cells have evolved multiple pathways to repair DNA double strand breaks (DSBs) and ensure genome stability. In addition to non-homologous end-joining (NHEJ) and homologous recombination (HR), cells evolved an error-prone repair pathway termed microhomology-mediated end joining (MMEJ). The mutagenic outcome of MMEJ derives from the activity of DNA polymerase theta (Polθ) - a multidomain enzyme that is minimally expressed in normal tissue but overexpressed in tumors. Polθ expression is particularly crucial for the proliferation of HR deficient cancer cells. As a result, this mutagenic repair emerged as an attractive target for cancer therapy, and inhibitors are currently in pre-clinical development. Here, we review the multifunctionality of this enigmatic polymerase, focusing on its role during DSB repair in mammalian cells and its impact on cancer genomes.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures

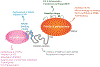
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

