The mitochondrial outer membrane protein SYNJ2BP interacts with the cell adhesion molecule TMIGD1 and can recruit it to mitochondria
- PMID: 32303178
- PMCID: PMC7164261
- DOI: 10.1186/s12860-020-00274-1
The mitochondrial outer membrane protein SYNJ2BP interacts with the cell adhesion molecule TMIGD1 and can recruit it to mitochondria
Abstract
Background: Transmembrane and immunoglobulin domain-containing protein 1 (TMIGD1) is a recently identified cell adhesion molecule which is predominantly expressed by epithelial cells of the intestine and the kidney. Its expression is downregulated in both colon and renal cancer suggesting a tumor suppressive activity. The function of TMIGD1 at the cellular level is largely unclear. Published work suggests a protective role of TMIGD1 during oxidative stress in kidney epithelial cells, but the underlying molecular mechanisms are unknown.
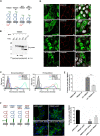
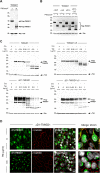
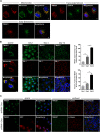
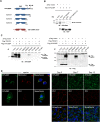
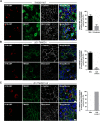
Results: In this study, we address the subcellular localization of TMIGD1 in renal epithelial cells and identify a cytoplasmic scaffold protein as interaction partner of TMIGD1. We find that TMIGD1 localizes to different compartments in renal epithelial cells and that this localization is regulated by cell confluency. Whereas it localizes to mitochondria in subconfluent cells it is localized at cell-cell contacts in confluent cells. We find that cell-cell contact localization is regulated by N-glycosylation and that both the extracellular and the cytoplasmic domain contribute to this localization. We identify Synaptojanin 2-binding protein (SYNJ2BP), a PDZ domain-containing cytoplasmic protein, which localizes to both mitochondria and the plasma membrane, as interaction partner of TMIGD1. The interaction of TMIGD1 and SYNJ2BP is mediated by the PDZ domain of SYNJ2BP and the C-terminal PDZ domain-binding motif of TMIGD1. We also find that SYNJ2BP can actively recruit TMIGD1 to mitochondria providing a potential mechanism for the localization of TMIGD1 at mitochondria.
Conclusions: This study describes TMIGD1 as an adhesion receptor that can localize to both mitochondria and cell-cell junctions in renal epithelial cells. It identifies SYNJ2BP as an interaction partner of TMIGD1 providing a potential mechanism underlying the localization of TMIGD1 at mitochondria. The study thus lays the basis for a better understanding of the molecular function of TMIGD1 during oxidative stress regulation.
Keywords: Adhesion molecule; Cell-cell adhesion; JAM; Kidney epithelium; SYNJ2BP; TMIGD1.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
TMIGD1 Inhibited Abdominal Adhesion Formation by Alleviating Oxidative Stress in the Mitochondria of Peritoneal Mesothelial Cells.Oxid Med Cell Longev. 2021 Aug 14;2021:9993704. doi: 10.1155/2021/9993704. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34426761 Free PMC article.
-
TMIGD1: Emerging functions of a tumor supressor and adhesion receptor.Oncogene. 2023 Jun;42(22):1777-1785. doi: 10.1038/s41388-023-02696-5. Epub 2023 Apr 22. Oncogene. 2023. PMID: 37087524 Free PMC article. Review.
-
TMIGD1 is a novel adhesion molecule that protects epithelial cells from oxidative cell injury.Am J Pathol. 2015 Oct;185(10):2757-67. doi: 10.1016/j.ajpath.2015.06.006. Epub 2015 Sep 2. Am J Pathol. 2015. PMID: 26342724 Free PMC article.
-
Membrane recruitment of the polarity protein Scribble by the cell adhesion receptor TMIGD1.Commun Biol. 2023 Jul 10;6(1):702. doi: 10.1038/s42003-023-05088-3. Commun Biol. 2023. PMID: 37430142 Free PMC article.
-
Physiological functions of junctional adhesion molecules (JAMs) in tight junctions.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183299. doi: 10.1016/j.bbamem.2020.183299. Epub 2020 Apr 2. Biochim Biophys Acta Biomembr. 2020. PMID: 32247783 Review.
Cited by
-
Longitudinal in vivo metabolic labeling reveals tissue-specific mitochondrial proteome turnover rates and proteins selectively altered by parkin deficiency.Sci Rep. 2023 Jul 14;13(1):11414. doi: 10.1038/s41598-023-38484-0. Sci Rep. 2023. PMID: 37452120 Free PMC article.
-
TMIGD1 Inhibited Abdominal Adhesion Formation by Alleviating Oxidative Stress in the Mitochondria of Peritoneal Mesothelial Cells.Oxid Med Cell Longev. 2021 Aug 14;2021:9993704. doi: 10.1155/2021/9993704. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34426761 Free PMC article.
-
Transcriptional sequencing analysis reveals the potential use of deer antler for "tonifying the kidney and strengthening bone".J Orthop Surg Res. 2022 Sep 14;17(1):419. doi: 10.1186/s13018-022-03308-w. J Orthop Surg Res. 2022. PMID: 36104709 Free PMC article.
-
TMIGD1: Emerging functions of a tumor supressor and adhesion receptor.Oncogene. 2023 Jun;42(22):1777-1785. doi: 10.1038/s41388-023-02696-5. Epub 2023 Apr 22. Oncogene. 2023. PMID: 37087524 Free PMC article. Review.
-
The Reciprocal Relationship Between Cell Adhesion Molecules and Reactive Oxygen Species.Cells. 2025 Jul 17;14(14):1098. doi: 10.3390/cells14141098. Cells. 2025. PMID: 40710351 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

