Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2
- PMID: 32303592
- PMCID: PMC7307149
- DOI: 10.1128/JVI.00510-20
Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2
Abstract
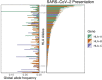
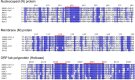
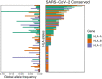
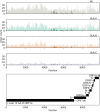
Genetic variability across the three major histocompatibility complex (MHC) class I genes (human leukocyte antigen A [HLA-A], -B, and -C genes) may affect susceptibility to and severity of the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for coronavirus disease 2019 (COVID-19). We performed a comprehensive in silico analysis of viral peptide-MHC class I binding affinity across 145 HLA-A, -B, and -C genotypes for all SARS-CoV-2 peptides. We further explored the potential for cross-protective immunity conferred by prior exposure to four common human coronaviruses. The SARS-CoV-2 proteome was successfully sampled and was represented by a diversity of HLA alleles. However, we found that HLA-B*46:01 had the fewest predicted binding peptides for SARS-CoV-2, suggesting that individuals with this allele may be particularly vulnerable to COVID-19, as they were previously shown to be for SARS (M. Lin, H.-T. Tseng, J. A. Trejaut, H.-L. Lee, et al., BMC Med Genet 4:9, 2003, https://bmcmedgenet.biomedcentral.com/articles/10.1186/1471-2350-4-9). Conversely, we found that HLA-B*15:03 showed the greatest capacity to present highly conserved SARS-CoV-2 peptides that are shared among common human coronaviruses, suggesting that it could enable cross-protective T-cell-based immunity. Finally, we reported global distributions of HLA types with potential epidemiological ramifications in the setting of the current pandemic.IMPORTANCE Individual genetic variation may help to explain different immune responses to a virus across a population. In particular, understanding how variation in HLA may affect the course of COVID-19 could help identify individuals at higher risk from the disease. HLA typing can be fast and inexpensive. Pairing HLA typing with COVID-19 testing where feasible could improve assessment of severity of viral disease in the population. Following the development of a vaccine against SARS-CoV-2, the virus that causes COVID-19, individuals with high-risk HLA types could be prioritized for vaccination.
Keywords: COVID-19; HLA; MHC class I; SARS-CoV-2; coronavirus.
Copyright © 2020 Nguyen et al.
Figures
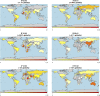
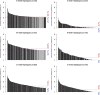
Comment in
-
Coronavirus Disease (Covid-19): What Are We Learning in a Country With High Mortality Rate?Front Immunol. 2020 May 28;11:1208. doi: 10.3389/fimmu.2020.01208. eCollection 2020. Front Immunol. 2020. PMID: 32574270 Free PMC article. No abstract available.
References
-
- World Health Organization. 2020. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). WHO (World Health Organization), Geneva, Switzerland.
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Roser M, Ritchie HR, Ortiz-Ospina E, Hasell J. 2020. Coronavirus disease (COVID-19)—statistics and research. https://ourworldindata.org/coronavirus.
-
- Caramelo F, Ferreira N, Oliveiros B. 2020. Estimation of risk factors for COVID-19 mortality-preliminary results. medRxiv doi: 10.1101/2020.02.24.20027268. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

