Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans
- PMID: 32303790
- PMCID: PMC11105112
- DOI: 10.1007/s00018-020-03518-7
Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans
Abstract
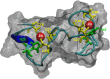
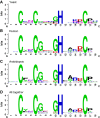
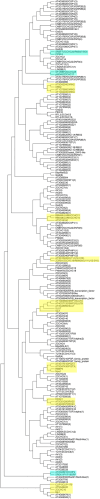
The diverse eukaryotic proteins that contain zinc fingers participate in many aspects of nucleic acid metabolism, from DNA transcription to RNA degradation, post-transcriptional gene silencing, and small RNA biogenesis. These proteins can be classified into at least 30 types based on structure. In this review, we focus on the CCHC-type zinc fingers (ZCCHC), which contain an 18-residue domain with the CX2CX4HX4C sequence, where C is cysteine, H is histidine, and X is any amino acid. This motif, also named the "zinc knuckle", is characteristic of the retroviral Group Antigen protein and occurs alone or with other motifs. Many proteins containing zinc knuckles have been identified in eukaryotes, but only a few have been studied. Here, we review the available information on ZCCHC-containing factors from three evolutionarily distant eukaryotes-Saccharomyces cerevisiae, Arabidopsis thaliana, and Homo sapiens-representing fungi, plants, and metazoans, respectively. We performed systematic searches for proteins containing the CX2CX4HX4C sequence in organism-specific and generalist databases. Next, we analyzed the structural and functional information for all such proteins stored in UniProtKB. Excluding retrotransposon-encoded proteins and proteins harboring uncertain ZCCHC motifs, we found seven ZCCHC-containing proteins in yeast, 69 in Arabidopsis, and 34 in humans. ZCCHC-containing proteins mainly localize to the nucleus, but some are nuclear and cytoplasmic, or exclusively cytoplasmic, and one localizes to the chloroplast. Most of these factors participate in RNA metabolism, including transcriptional elongation, polyadenylation, translation, pre-messenger RNA splicing, RNA export, RNA degradation, microRNA and ribosomal RNA biogenesis, and post-transcriptional gene silencing. Several human ZCCHC-containing factors are derived from neofunctionalized retrotransposons and act as proto-oncogenes in diverse neoplastic processes. The conservation of ZCCHCs in orthologs of these three phylogenetically distant eukaryotes suggests that these domains have biologically relevant functions that are not well known at present.
Keywords: Arabidopsis; Human; RNA metabolism; Yeast; ZCCHC; Zinc-knuckle.
Figures



References
-
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

