Tapping into the maize root microbiome to identify bacteria that promote growth under chilling conditions
- PMID: 32305066
- PMCID: PMC7166315
- DOI: 10.1186/s40168-020-00833-w
Tapping into the maize root microbiome to identify bacteria that promote growth under chilling conditions
Abstract
Background: When maize (Zea mays L.) is grown in the Northern hemisphere, its development is heavily arrested by chilling temperatures, especially at the juvenile phase. As some endophytes are beneficial for plants under stress conditions, we analyzed the impact of chilling temperatures on the root microbiome and examined whether microbiome-based analysis might help to identify bacterial strains that could promote growth under these temperatures.
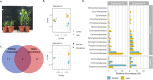
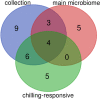
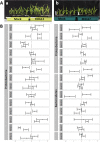
Results: We investigated how the maize root microbiome composition changed by means of 16S rRNA gene amplicon sequencing when maize was grown at chilling temperatures in comparison to ambient temperatures by repeatedly cultivating maize in field soil. We identified 12 abundant and enriched bacterial families that colonize maize roots, consisting of bacteria recruited from the soil, whereas seed-derived endophytes were lowly represented. Chilling temperatures modified the root microbiome composition only slightly, but significantly. An enrichment of several chilling-responsive families was detected, of which the Comamonadaceae and the Pseudomonadaceae were the most abundant in the root endosphere of maize grown under chilling conditions, whereas only three were strongly depleted, among which the Streptomycetaceae. Additionally, a collection of bacterial strains isolated from maize roots was established and a selection was screened for growth-promoting effects on juvenile maize grown under chilling temperatures. Two promising strains that promoted maize growth under chilling conditions were identified that belonged to the root endophytic bacterial families, from which the relative abundance remained unchanged by variations in the growth temperature.
Conclusions: Our analyses indicate that chilling temperatures affect the bacterial community composition within the maize root endosphere. We further identified two bacterial strains that boost maize growth under chilling conditions. Their identity revealed that analyzing the chilling-responsive families did not help for their identification. As both strains belong to root endosphere enriched families, visualizing and comparing the bacterial diversity in these communities might still help to identify new PGPR strains. Additionally, a strain does not necessarely need to belong to a high abundant family in the root endosphere to provoke a growth-promoting effect in chilling conditions. Video abstract.
Keywords: Chilling temperatures; Maize; Microbiome; PGPR; Plant growth-promoting rhizobacteria; Root endosphere.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Farooq M, Aziz T, Wahid A, Lee D-J, Siddique KHM. Chilling tolerance in maize: agronomic and physiological approaches. Crop Pasture Sci. 2009;60:501–516. doi: 10.1071/CP08427. - DOI
-
- Leipner J, Stamp P. Chilling stress in maize seedlings. In: Bennetzen JL, Hake SC, editors. Handbook of Maize: Its Biology. New York, NY: Springer; 2009. pp. 291–310.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

