Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2
- PMID: 32306853
- PMCID: PMC7301696
- DOI: 10.1080/22221751.2020.1756698
Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2
Abstract
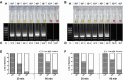
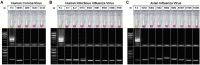
The previous outbreaks of SARS-CoV and MERS-CoV have led researchers to study the role of diagnostics in impediment of further spread and transmission. With the recent emergence of the novel SARS-CoV-2, the availability of rapid, sensitive, and reliable diagnostic methods is essential for disease control. Hence, we have developed a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the specific detection of SARS-CoV-2. The primer sets for RT-LAMP assay were designed to target the nucleocapsid gene of the viral RNA, and displayed a detection limit of 102 RNA copies close to that of qRT-PCR
Keywords: COVID-19; SARS-CoV-2; colorimetric detection; molecular diagnosis; novel coronavirus; reverse transcription-loop-mediated isothermal amplification.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
