Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes
- PMID: 32306862
- PMCID: PMC7189413
- DOI: 10.1080/07391102.2020.1758791
Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes
Abstract
In December 2019, COVID-19 epidemic was described in Wuhan, China, and the infection has spread widely affecting hundreds of thousands. Herein, an effort was made to identify commercially available drugs in order to repurpose them against coronavirus by the means of structure-based virtual screening. In addition, ZINC15 library was used to identify novel leads against main proteases. Human TMPRSS2 3D structure was first generated using homology modeling approach. Our molecular docking study showed four potential inhibitors against Mpro enzyme, two available drugs (Talampicillin and Lurasidone) and two novel drug-like compounds (ZINC000000702323 and ZINC000012481889). Moreover, four promising inhibitors were identified against TMPRSS2; Rubitecan and Loprazolam drugs, and compounds ZINC000015988935 and ZINC000103558522. ADMET profile showed that the hits from our study are safe and drug-like compounds. Furthermore, molecular dynamic (MD) simulation and binding free energy calculation using the MM-PBSA method was performed to calculate the interaction energy of the top-ranked drugs.Communicated by Ramaswamy H. Sarma.
Keywords: ADMET; COVID-19; MD simulation; MM-PBSA; homology modeling; structure-based virtual screening.
Figures
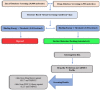


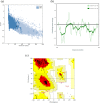

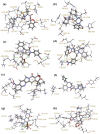

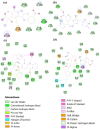

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
