On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments
- PMID: 32306892
- PMCID: PMC7168886
- DOI: 10.1186/s12864-020-6721-y
On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments
Erratum in
-
Correction to: On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments.BMC Genomics. 2020 Jun 3;21(1):384. doi: 10.1186/s12864-020-6754-2. BMC Genomics. 2020. PMID: 32493350 Free PMC article.
Abstract
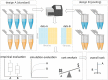
Background: In gene expression studies, RNA sample pooling is sometimes considered because of budget constraints or lack of sufficient input material. Using microarray technology, RNA sample pooling strategies have been reported to optimize both the cost of data generation as well as the statistical power for differential gene expression (DGE) analysis. For RNA sequencing, with its different quantitative output in terms of counts and tunable dynamic range, the adequacy and empirical validation of RNA sample pooling strategies have not yet been evaluated. In this study, we comprehensively assessed the utility of pooling strategies in RNA-seq experiments using empirical and simulated RNA-seq datasets.
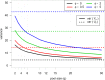
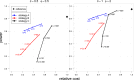
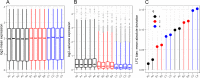
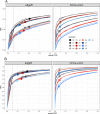
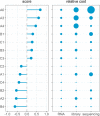
Result: The data generating model in pooled experiments is defined mathematically to evaluate the mean and variability of gene expression estimates. The model is further used to examine the trade-off between the statistical power of testing for DGE and the data generating costs. Empirical assessment of pooling strategies is done through analysis of RNA-seq datasets under various pooling and non-pooling experimental settings. Simulation study is also used to rank experimental scenarios with respect to the rate of false and true discoveries in DGE analysis. The results demonstrate that pooling strategies in RNA-seq studies can be both cost-effective and powerful when the number of pools, pool size and sequencing depth are optimally defined.
Conclusion: For high within-group gene expression variability, small RNA sample pools are effective to reduce the variability and compensate for the loss of the number of replicates. Unlike the typical cost-saving strategies, such as reducing sequencing depth or number of RNA samples (replicates), an adequate pooling strategy is effective in maintaining the power of testing DGE for genes with low to medium abundance levels, along with a substantial reduction of the total cost of the experiment. In general, pooling RNA samples or pooling RNA samples in conjunction with moderate reduction of the sequencing depth can be good options to optimize the cost and maintain the power.
Keywords: Cost; Differential gene expression; Experimental design; RNA sample pooling; RNA sequencing; Statistical power.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

