Epigenomic Assessment of Cardiovascular Disease Risk and Interactions With Traditional Risk Metrics
- PMID: 32308120
- PMCID: PMC7428544
- DOI: 10.1161/JAHA.119.015299
Epigenomic Assessment of Cardiovascular Disease Risk and Interactions With Traditional Risk Metrics
Abstract
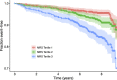
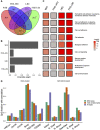
Background Epigenome-wide association studies for cardiometabolic risk factors have discovered multiple loci associated with incident cardiovascular disease (CVD). However, few studies have sought to directly optimize a predictor of CVD risk. Furthermore, it is challenging to train multivariate models across multiple studies in the presence of study- or batch effects. Methods and Results Here, we analyzed existing DNA methylation data collected using the Illumina HumanMethylation450 microarray to create a predictor of CVD risk across 3 cohorts: Women's Health Initiative, Framingham Heart Study Offspring Cohort, and Lothian Birth Cohorts. We trained Cox proportional hazards-based elastic net regressions for incident CVD separately in each cohort and used a recently introduced cross-study learning approach to integrate these individual scores into an ensemble predictor. The methylation-based risk score was associated with CVD time-to-event in a held-out fraction of the Framingham data set (hazard ratio per SD=1.28, 95% CI, 1.10-1.50) and predicted myocardial infarction status in the independent REGICOR (Girona Heart Registry) data set (odds ratio per SD=2.14, 95% CI, 1.58-2.89). These associations remained after adjustment for traditional cardiovascular risk factors and were similar to those from elastic net models trained on a directly merged data set. Additionally, we investigated interactions between the methylation-based risk score and both genetic and biochemical CVD risk, showing preliminary evidence of an enhanced performance in those with less traditional risk factor elevation. Conclusions This investigation provides proof-of-concept for a genome-wide, CVD-specific epigenomic risk score and suggests that DNA methylation data may enable the discovery of high-risk individuals who would be missed by alternative risk metrics.
Keywords: DNA methylation; cardiovascular disease; epigenomics; risk prediction.
Figures


References
-
- Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, van Iterson M, van Dijk F, van Galen M, Bot J, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2016;49:131–138. - PubMed
Publication types
MeSH terms
Grants and funding
- HHSN268201600001C/HL/NHLBI NIH HHS/United States
- HHSN268201600003C/HL/NHLBI NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- MR/M01311/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201600002C/HL/NHLBI NIH HHS/United States
- HHSN268201600018C/HL/NHLBI NIH HHS/United States
- G0700704/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- T32 HL069772/HL/NHLBI NIH HHS/United States
- HHSN268201600004C/HL/NHLBI NIH HHS/United States
- N01 HC025195/HC/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

