Starvation-induced suppression of DAZAP1 by miR-10b integrates splicing control into TSC2-regulated oncogenic autophagy in esophageal squamous cell carcinoma
- PMID: 32308763
- PMCID: PMC7163442
- DOI: 10.7150/thno.43046
Starvation-induced suppression of DAZAP1 by miR-10b integrates splicing control into TSC2-regulated oncogenic autophagy in esophageal squamous cell carcinoma
Abstract
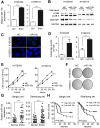
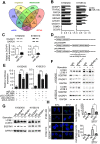
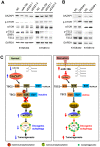
Esophageal squamous cell carcinoma (ESCC) accounts for about 90% of all incident esophageal cancers, with a 5-year survival rate of < 20%. Autophagy is of particular importance in cancers; however, the detailed regulatory mechanisms of oncogenic autophagy in ESCC have not been fully elucidated. In the present study, we address how splicing control of TSC2 is involved in mTOR-regulated oncogenic autophagy. Methods: Alternative splicing events controlled by DAZAP1 in ESCC cells were identified via RNAseq. Differential phosphorylation of short or long TSC2 splicing variants by AKT and their impacts on mTOR signaling were also examined. Results: We found that starvation-induced miR-10b could enhance autophagy via silencing DAZAP1, a key regulator of pre-mRNA alternative splicing. Intriguingly, we observed a large number of significantly changed alternative splicing events, especially exon skipping, upon RNAi of DAZAP1. TSC2 was verified as one of the crucial target genes of DAZAP1. Silencing of DAZAP1 led to the exclusion of TSC2 exon 26 (from Leu947 to Arg988), producing a short TSC2 isoform. The short TSC2 isoform cannot be phosphorylated at Ser981 by AKT, which resulted in continuous activation of TSC2 in ESCC. The active TSC2 inhibited mTOR via RHEB, leading to continually stimulated oncogenic autophagy of ESCC cells. Conclusions: Our data revealed an important physiological function of tumor suppressor DAZAP1 in autophagy regulation and highlighted the potential of controlling mRNA alternative splicing as an effective therapeutic application for cancers.
Keywords: DAZAP1; TSC2; autophagy; esophageal squamous cell carcinoma; miR-10b; selective splicing.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

