Screening for Novel LOX and SOD1 Variants in Keratoconus Patients from Brazil
- PMID: 32308947
- PMCID: PMC7151510
- DOI: 10.18502/jovr.v15i2.6730
Screening for Novel LOX and SOD1 Variants in Keratoconus Patients from Brazil
Abstract
Purpose: To investigate the presence of the variants of lysyl oxygenase (LOX) and superoxide dismutase 1 (SOD1) genes in Brazilian patients with advanced keratoconus.
Methods: Donor genomic DNA extracted from blood samples was screened for 5'UTR, exonic LOX, and SOD1 variants in a subset of 26 patients presenting with advanced keratoconus (KISA 1000% and I-S 2.0) by Sanger sequencing. The impact of non-synonymous amino acid changes was evaluated by SIFT, PMUT, and PolyPhen algorithms. The Mutation Taster tool was used to evaluate the potential impact of formation of new donor and acceptor splice sites in the promoter region of affected volunteers carrying sequence variants. A 7-base SOD1 deletion (IVS2 + 50del7bp) previously associated with keratoconus was screened in 140 patients presenting classical keratoconus by gel fragment analysis, and positive samples were sequenced for confirmation.
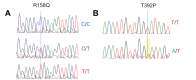
Results: We found an unreported missense variant in LOX exon 6 in one heterozygous patient, leading to substitution of proline with threonine at residue 392 (p. Thr392Pro) of LOX protein sequence. This mutation was predicted to be potentially damaging to LOX protein. Another LOX variant, Arg158Gln, was also detected in another patient but predicted to be non-pathogenic. Two additional new polymorphisms in LOX 5'UTR region (-116C T and -58C T) were found in two patients presenting with advanced keratoconus and were predicted to modulate or create donor/acceptor splice sites in LOX transcripts. Additionally, SOD1 deletion was detected in one patient presenting with severe keratoconus, not in control samples.
Conclusion: We described three novel LOX polymorphisms identified for the first time in Brazilian patients with advanced keratoconus, as well as a previously described SOD1 deletion strongly associated with keratoconus. A possible role of these variants in modulating transcript levels in the cornea of affected individual requires further investigation.
Keywords: LOX; Mutation; SOD1; Keratoconus.
Copyright © 2020 Gadelha et al.
Conflict of interest statement
There are no conflicts of interest.
Figures



References
-
- Abto Brazilian Association of Organ and Tissue Transplantation. Report on performed transplants - Jan/Dec 2018 [Internet]; 2019. Available from: abto.org.br .
-
- J Lechner, Ha Bae, J Guduric-Fuchs, A Rice, G Govindarajan, S Siddiqui, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci 2013;54:5266–5272. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
