RA-map: building a state-of-the-art interactive knowledge base for rheumatoid arthritis
- PMID: 32311035
- PMCID: PMC7170216
- DOI: 10.1093/database/baaa017
RA-map: building a state-of-the-art interactive knowledge base for rheumatoid arthritis
Abstract
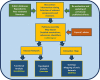
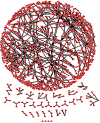
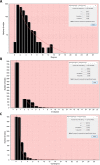
Rheumatoid arthritis (RA) is a progressive, inflammatory autoimmune disease of unknown aetiology. The complex mechanism of aetiopathogenesis, progress and chronicity of the disease involves genetic, epigenetic and environmental factors. To understand the molecular mechanisms underlying disease phenotypes, one has to place implicated factors in their functional context. However, integration and organization of such data in a systematic manner remains a challenging task. Molecular maps are widely used in biology to provide a useful and intuitive way of depicting a variety of biological processes and disease mechanisms. Recent large-scale collaborative efforts such as the Disease Maps Project demonstrate the utility of such maps as versatile tools to organize and formalize disease-specific knowledge in a comprehensive way, both human and machine-readable. We present a systematic effort to construct a fully annotated, expert validated, state-of-the-art knowledge base for RA in the form of a molecular map. The RA map illustrates molecular and signalling pathways implicated in the disease. Signal transduction is depicted from receptors to the nucleus using the Systems Biology Graphical Notation (SBGN) standard representation. High-quality manual curation, use of only human-specific studies and focus on small-scale experiments aim to limit false positives in the map. The state-of-the-art molecular map for RA, using information from 353 peer-reviewed scientific publications, comprises 506 species, 446 reactions and 8 phenotypes. The species in the map are classified to 303 proteins, 61 complexes, 106 genes, 106 RNA entities, 2 ions and 7 simple molecules. The RA map is available online at ramap.elixir-luxembourg.org as an open-access knowledge base allowing for easy navigation and search of molecular pathways implicated in the disease. Furthermore, the RA map can serve as a template for omics data visualization.
© The Author(s) 2020. Published by Oxford University Press.
Figures



References
-
- McInnes I.B. and Schett G. (2011) The pathogenesis of rheumatoid arthritis. N. Engl. J. Med., 365, 2205–2219. - PubMed
-
- Frank-Bertoncelj M., Klein K. and Gay S. (2017) Interplay between genetic and epigenetic mechanisms in rheumatoid arthritis. Epigenomics, 9, 493–504. - PubMed
-
- Niarakis A., Bounab Y., Grieco L. et al. (2014) Computational modeling of the main signaling pathways involved in mast cell activation. Curr. Top. Microbiol. Immunol., 382, 69–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

