Tumor-associated macrophages promote ovarian cancer cell migration by secreting transforming growth factor beta induced (TGFBI) and tenascin C
- PMID: 32312959
- PMCID: PMC7171168
- DOI: 10.1038/s41419-020-2438-8
Tumor-associated macrophages promote ovarian cancer cell migration by secreting transforming growth factor beta induced (TGFBI) and tenascin C
Abstract
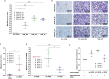
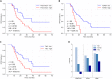
A central and unique aspect of high-grade serous ovarian carcinoma (HGSC) is the extensive transcoelomic spreading of tumor cell via the peritoneal fluid or malignant ascites. We and others identified tumor-associated macrophages (TAM) in the ascites as promoters of metastasis-associated processes like extracellular matrix (ECM) remodeling, tumor cell migration, adhesion, and invasion. The precise mechanisms and mediators involved in these functions of TAM are, however, largely unknown. We observed that HGSC migration is promoted by soluble mediators from ascites-derived TAM, which can be emulated by conditioned medium from monocyte-derived macrophages (MDM) differentiated in ascites to TAM-like asc-MDM. A similar effect was observed with IL-10-induced alternatively activated m2c-MDM but not with LPS/IFNγ-induced inflammatory m1-MDM. These observations provided the basis for deconvolution of the complex TAM secretome by performing comparative secretome analysis of matched triplets of different MDM phenotypes with different pro-migratory properties (asc-MDM, m2c-MDM, m1-MDM). Mass spectrometric analysis identified an overlapping set of nine proteins secreted by both asc-MDM and m2c-MDM, but not by m1-MDM. Of these, three proteins, i.e., transforming growth factor beta-induced (TGFBI) protein, tenascin C (TNC), and fibronectin (FN1), have been associated with migration-related functions. Intriguingly, increased ascites concentrations of TGFBI, TNC, and fibronectin were associated with short progression-free survival. Furthermore, transcriptome and secretome analyses point to TAM as major producers of these proteins, further supporting an essential role for TAM in promoting HGSC progression. Consistent with this hypothesis, we were able to demonstrate that the migration-inducing potential of asc-MDM and m2c-MDM secretomes is inhibited, at least partially, by neutralizing antibodies against TGFBI and TNC or siRNA-mediated silencing of TGFBI expression. In conclusion, the present study provides the first experimental evidence that TAM-derived TGFBI and TNC in ascites promote HGSC progression.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

