High throughput pMHC-I tetramer library production using chaperone-mediated peptide exchange
- PMID: 32312993
- PMCID: PMC7170893
- DOI: 10.1038/s41467-020-15710-1
High throughput pMHC-I tetramer library production using chaperone-mediated peptide exchange
Abstract
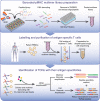
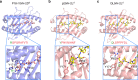
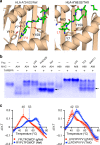
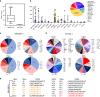
Peptide exchange technologies are essential for the generation of pMHC-multimer libraries used to probe diverse, polyclonal TCR repertoires in various settings. Here, using the molecular chaperone TAPBPR, we develop a robust method for the capture of stable, empty MHC-I molecules comprising murine H2 and human HLA alleles, which can be readily tetramerized and loaded with peptides of choice in a high-throughput manner. Alternatively, catalytic amounts of TAPBPR can be used to exchange placeholder peptides with high affinity peptides of interest. Using the same system, we describe high throughput assays to validate binding of multiple candidate peptides on empty MHC-I/TAPBPR complexes. Combined with tetramer-barcoding via a multi-modal cellular indexing technology, ECCITE-seq, our approach allows a combined analysis of TCR repertoires and other T cell transcription profiles together with their cognate antigen specificities in a single experiment. The new approach allows TCR/pMHC interactions to be interrogated easily at large scale.
Conflict of interest statement
N.G.S. and P.S. are listed as inventor and co-inventor on patent applications related to the preparation of peptide-deficient MHC-I/Chaperone complexes and ECCITE-seq, respectively (US patent number WO 2020/010261 and provisional application 62/694-824). The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

