A Phosphorylated Intermediate in the Activation of WNK Kinases
- PMID: 32314908
- PMCID: PMC7914002
- DOI: 10.1021/acs.biochem.0c00146
A Phosphorylated Intermediate in the Activation of WNK Kinases
Abstract
WNK kinases autoactivate by autophosphorylation. Crystallography of the kinase domain of WNK1 phosphorylated on the primary activating site (pWNK1) in the presence of AMP-PNP reveals a well-ordered but inactive configuration. This new pWNK1 structure features specific and unique interactions of the phosphoserine, less hydration, and smaller cavities compared with those of unphosphorylated WNK1 (uWNK1). Because WNKs are activated by osmotic stress in cells, we addressed whether the structure was influenced directly by osmotic pressure. pWNK1 crystals formed in PEG3350 were soaked in the osmolyte sucrose. Suc-WNK1 crystals maintained X-ray diffraction, but the lattice constants and pWNK1 structure changed. Differences were found in the activation loop and helix C, common switch loci in kinase activation. On the basis of these structural changes, we tested for effects on in vitro activity of two WNKs, pWNK1 and pWNK3. The osmolyte PEG400 enhanced ATPase activity. Our data suggest multistage activation of WNKs.
Figures
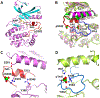
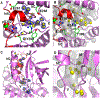


References
-
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, and Cobb MH (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II, J Biol Chem 275, 16795–16801. - PubMed
-
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, and Lifton RP (2001) Human hypertension caused by mutations in WNK kinases, Science 293, 1107–1112. - PubMed
-
- Lytle C, and Forbush B 3rd. (1996) Regulatory phosphorylation of the secretory Na-K-Cl cotransporter: modulation by cytoplasmic Cl, Am J Physiol 270, C437–448. - PubMed
-
- Lytle C, and Forbush B 3rd. (1992) Na-K-Cl cotransport in the shark rectal gland. II. Regulation in isolated tubules, Am J Physiol 262, C1009–1017. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

