MiR193a Modulation and Podocyte Phenotype
- PMID: 32316697
- PMCID: PMC7226544
- DOI: 10.3390/cells9041004
MiR193a Modulation and Podocyte Phenotype
Abstract
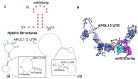
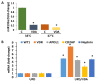
Apolipoprotein L1 (APOL1)-miR193a axis has been reported to play a role in the maintenance of podocyte homeostasis. In the present study, we analyzed transcription factors relevant to miR193a in human podocytes and their effects on podocytes' molecular phenotype. The motif scan of the miR193a gene provided information about transcription factors, including YY1, WT1, Sox2, and VDR-RXR heterodimer, which could potentially bind to the miR193a promoter region to regulate miR193a expression. All structure models of these transcription factors and the tertiary structures of the miR193a promoter region were generated and refined using computational tools. The DNA-protein complexes of the miR193a promoter region and transcription factors were created using a docking approach. To determine the modulatory role of miR193a on APOL1 mRNA, the structural components of APOL1 3' UTR and miR193a-5p were studied. Molecular Dynamic (MD) simulations validated interactions between miR193a and YY1/WT1/Sox2/VDR/APOL1 3' UTR region. Undifferentiated podocytes (UPDs) displayed enhanced miR193a, YY1, and Sox2 but attenuated WT1, VDR, and APOL1 expressions, whereas differentiated podocytes (DPDs) exhibited attenuated miR193a, YY1, and Sox2 but increased WT1, VDR, APOL1 expressions. Inhibition of miR193a in UPDs enhanced the expression of APOL1 as well as of podocyte molecular markers; on the other hand, DPD-transfected with miR193a plasmid showed downing of APOL1 as well as podocyte molecular markers suggesting a causal relationship between miR193a and podocyte molecular markers. Silencing of YY1 and Sox2 in UPDs decreased the expression of miR193a but increased the expression of VDR, and CD2AP (a marker of DPDs); in contrast, silencing of WT1 and VDR in DPDs enhanced the expression of miR193a, YY1, and Sox2. Since miR193a-downing by Vitamin D receptor (VDR) agonist not only enhanced the mRNA expression of APOL1 but also of podocyte differentiating markers, suggest that down-regulation of miR193a could be used to enhance the expression of podocyte differentiating markers as a therapeutic strategy.
Keywords: APOL1; RXR; Sox2; VDR; WT1; YY1; miR193a.
Conflict of interest statement
None of the authors have any conflicts of interest.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

