Circadian regulation of mitochondrial uncoupling and lifespan
- PMID: 32317636
- PMCID: PMC7174288
- DOI: 10.1038/s41467-020-15617-x
Circadian regulation of mitochondrial uncoupling and lifespan
Abstract
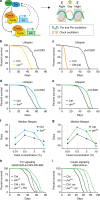
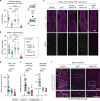
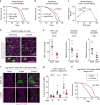
Because old age is associated with defects in circadian rhythm, loss of circadian regulation is thought to be pathogenic and contribute to mortality. We show instead that loss of specific circadian clock components Period (Per) and Timeless (Tim) in male Drosophila significantly extends lifespan. This lifespan extension is not mediated by canonical diet-restriction longevity pathways but is due to altered cellular respiration via increased mitochondrial uncoupling. Lifespan extension of per mutants depends on mitochondrial uncoupling in the intestine. Moreover, upregulated uncoupling protein UCP4C in intestinal stem cells and enteroblasts is sufficient to extend lifespan and preserve proliferative homeostasis in the gut with age. Consistent with inducing a metabolic state that prevents overproliferation, mitochondrial uncoupling drugs also extend lifespan and inhibit intestinal stem cell overproliferation due to aging or even tumorigenesis. These results demonstrate that circadian-regulated intestinal mitochondrial uncoupling controls longevity in Drosophila and suggest a new potential anti-aging therapeutic target.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM130764/GM/NIGMS NIH HHS/United States
- T32 HL120826/HL/NHLBI NIH HHS/United States
- R01 GM117407/GM/NIGMS NIH HHS/United States
- P40 OD018537/OD/NIH HHS/United States
- F31 GM125363/GM/NIGMS NIH HHS/United States
- R35 GM119793/GM/NIGMS NIH HHS/United States
- R21 DK112074/DK/NIDDK NIH HHS/United States
- R35 GM124717/GM/NIGMS NIH HHS/United States
- R01 AG045842/AG/NIA NIH HHS/United States
- R21 AR077312/AR/NIAMS NIH HHS/United States
- T32 DK007328/DK/NIDDK NIH HHS/United States
- R35 GM127049/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

